
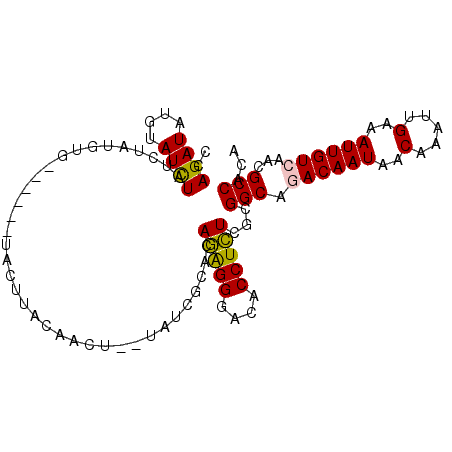
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,664,446 – 10,664,543 |
| Length | 97 |
| Max. P | 0.954327 |

| Location | 10,664,446 – 10,664,543 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10664446 97 + 22407834 CCAGAUAUGUAUCUAUCUAUGUG------UACUUACAACUCGUAUAGCACAGAGGGACACCUCUAGCGGCUGACAAUAACAAAUUGAAAUUGUCAACGCCACA ..(((((......)))))..(((------(...(((.....)))..))))(((((....))))).(.((((((((((..(.....)..)))))))..))).). ( -25.00) >DroSec_CAF1 17614 92 + 1 CCAGAUAUGUAUCUAUCUAUGUG------CACUUACAACUCGUAU-----AGAGGGACACCUCUCGUGGCUGACAAUAACAAAUUGAAAUUGUCAACGCCACA ..(((((......))))).((((------(...........))))-----)((((....))))..((((((((((((..(.....)..)))))))..))))). ( -24.30) >DroSim_CAF1 17574 97 + 1 CCAGAUAUGUAUCUAUCUAUGUG------CACUUACAACUCGUAUCGCACAGAGGGACACCUCUCGCGGCUGACAAUAACAAAUUGAAAUUGUCAACGCCACA ..(((((......))))).((((------(...(((.....)))..)))))((((....))))..(.((((((((((..(.....)..)))))))..))).). ( -26.50) >DroEre_CAF1 20035 90 + 1 CCAGAUUUGUAUCUAUCCAUGUG------UAUUCA-------UAUCGCACAGAGGGACACCUCUCGCGGCAGACAAUAACAAAUUGAAAUUGUCAACGCCACA ..((((....))))....(((((------....))-------))).((..(((((....))))).))(((.((((((..(.....)..))))))...)))... ( -20.60) >DroYak_CAF1 18629 98 + 1 CCAGAUAUGUAUCUAUGAAUGUAGCUAUGUAAGUAUC-----AACCACACAGAGGGACACCUCUCGCGGCAAACAAUAACAAAUUGAAAUUGUCAACGCCACA ...((((((((.((((....)))).)))....)))))-----........(((((....))))).(.(((..(((((..(.....)..)))))....))).). ( -17.40) >DroAna_CAF1 11892 87 + 1 CUAGAUACCAAUUUUUUUAUAAA------UA--AACGACU--------GCAAGGGAACACCUUUUACGGCAAACAAUAACAAAUUGAAAUUGUCAACGCCACA .......................------..--.......--------((((((.....))))....(((((.((((.....))))...)))))...)).... ( -9.20) >consensus CCAGAUAUGUAUCUAUCUAUGUG______UACUUACAACU__UAUCGCACAGAGGGACACCUCUCGCGGCAGACAAUAACAAAUUGAAAUUGUCAACGCCACA ..((((....))))....................................(((((....)))))...(((.((((((..(.....)..))))))...)))... (-14.30 = -14.08 + -0.22)


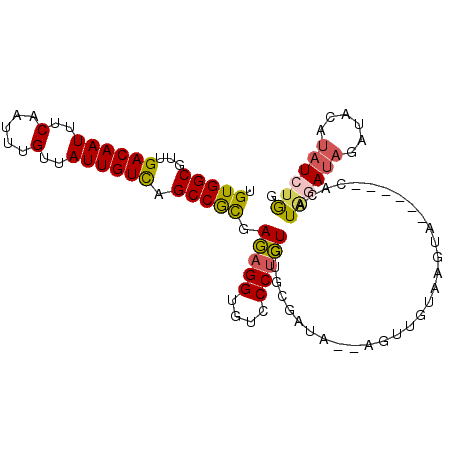
| Location | 10,664,446 – 10,664,543 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -19.96 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10664446 97 - 22407834 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUCAGCCGCUAGAGGUGUCCCUCUGUGCUAUACGAGUUGUAAGUA------CACAUAGAUAGAUACAUAUCUGG .(((((..(((((((..(.....)..)))))))))))).(((((....)))))(((((.(((.....)))))))------)...((((((......)))))). ( -30.70) >DroSec_CAF1 17614 92 - 1 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUCAGCCACGAGAGGUGUCCCUCU-----AUACGAGUUGUAAGUG------CACAUAGAUAGAUACAUAUCUGG ((((..(((((((((..(.....)..)))))))))(((.(((((....)))))-----.(((.....))).)))------))))((((((......)))))). ( -26.30) >DroSim_CAF1 17574 97 - 1 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUCAGCCGCGAGAGGUGUCCCUCUGUGCGAUACGAGUUGUAAGUG------CACAUAGAUAGAUACAUAUCUGG .(((((..(((((((..(.....)..)))))))))))).(((((....)))))(((((.(((.....)))..))------))).((((((......)))))). ( -32.50) >DroEre_CAF1 20035 90 - 1 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUCUGCCGCGAGAGGUGUCCCUCUGUGCGAUA-------UGAAUA------CACAUGGAUAGAUACAAAUCUGG .((((((..((((((..(.....)..)))))))))))).(((.(((((..((((((.(...-------......------).))))))..)))))...))).. ( -27.10) >DroYak_CAF1 18629 98 - 1 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUUUGCCGCGAGAGGUGUCCCUCUGUGUGGUU-----GAUACUUACAUAGCUACAUUCAUAGAUACAUAUCUGG (((((((((((.....))))).(((...((((.(((((((((((....))))).)))))).-----))))...)))..))))))....(((((....))))). ( -29.40) >DroAna_CAF1 11892 87 - 1 UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUUUGCCGUAAAAGGUGUUCCCUUGC--------AGUCGUU--UA------UUUAUAAAAAAAUUGGUAUCUAG ...((((..((((((..(.....)..))))))))))....((((.....)))).(--------(((..((--(.------......)))..))))........ ( -12.60) >consensus UGUGGCGUUGACAAUUUCAAUUUGUUAUUGUCAGCCGCGAGAGGUGUCCCUCUGUGCGAUA__AGUUGUAAGUA______CACAUAGAUAGAUACAUAUCUGG .(((((...((((((..(.....)..)))))).))))).(((((....)))))...............................((((((......)))))). (-19.96 = -19.93 + -0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:14 2006