| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,650,922 – 10,651,020 |
| Length | 98 |
| Max. P | 0.780508 |

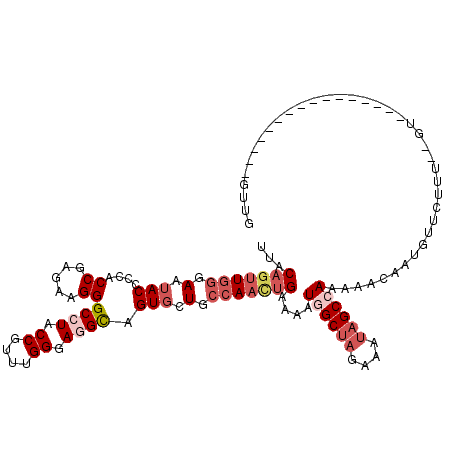
| Location | 10,650,922 – 10,651,020 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -18.10 |
| Energy contribution | -20.14 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10650922 98 + 22407834 --------------------CC--AAAGAGCAUAGUUUUUGGCUAAUUCUAGCCAUUUCCAAUUGGCAACACUGCCUCCCAAACGGUAGGCCCUUCUCGGUGUGGUAUUCCCAACUGUAA --------------------..--.......((((((..((((((....))))))..........((.(((((((((.((....)).)))).......))))).))......)))))).. ( -23.21) >DroSec_CAF1 4043 102 + 1 CAAC----------------AC--AAAGAACAUUGUUUUUGGCUUCUUAUAGCCAUUUUCAGUUGGCAGCACUGCCUGCCAAACGGUAGGCCCUUCUCGGUGGGGUAUUCCCAACUGUAA ((((----------------((--((......))))...(((((......)))))......))))(((((((((((((((....))))))).......))))(((....)))..)))).. ( -32.51) >DroSim_CAF1 4015 102 + 1 CAAC----------------AC--AAAGAACAUUGUUUUUGGCUAUUUAUAGCCAUUUUCAGUUGGCAGCACUGCCUGCCAAACGGUAGGCCCUUCUCGGUGGGGUAUUCCCAACUGUAA ((((----------------((--((......))))...((((((....))))))......))))(((((((((((((((....))))))).......))))(((....)))..)))).. ( -33.11) >DroEre_CAF1 6399 94 + 1 CAAC-------------------------ACCUCAG-UUUGGCUAUUUCUAGCGAUUUCCAGUUGGCAGCACUACAUCCCACACGGUAGGCCCUUCUCGGUGGGGUUUUCCCAUCUGUAA ...(-------------------------(((..((-...(((.....(((((........))))).....((((..........)))))))...)).))))(((....)))........ ( -20.60) >DroYak_CAF1 4726 119 + 1 AAAUAGUAUAAAUACCCAUUACAGAAUGAACGUAGU-UUUGGCAAUUUCUAGCGAUUUUCCCUUGCCAGCACUACUUCCCAUACGGUAGGCCCUUCUCGGUGGGGUAUUCCAAACUGUGA ..................((((((...(((.(((((-.(((((((.................))))))).))))))))......((...(((((.......)))))...))...)))))) ( -30.13) >consensus CAAC________________AC__AAAGAACAUAGUUUUUGGCUAUUUCUAGCCAUUUUCAGUUGGCAGCACUGCCUCCCAAACGGUAGGCCCUUCUCGGUGGGGUAUUCCCAACUGUAA .......................................((((((....))))))....(((((((.....(((((........)))))(((((.......)))))....)))))))... (-18.10 = -20.14 + 2.04)

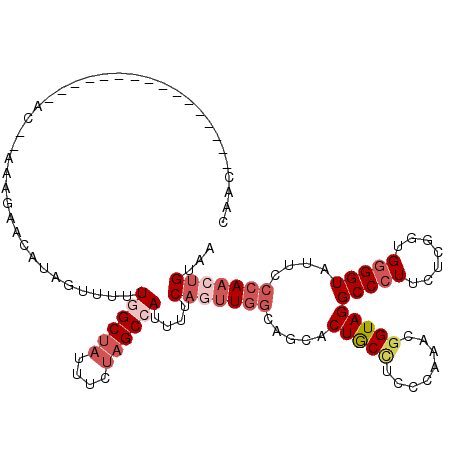
| Location | 10,650,922 – 10,651,020 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -19.36 |
| Energy contribution | -21.40 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10650922 98 - 22407834 UUACAGUUGGGAAUACCACACCGAGAAGGGCCUACCGUUUGGGAGGCAGUGUUGCCAAUUGGAAAUGGCUAGAAUUAGCCAAAAACUAUGCUCUUU--GG-------------------- ...(((((((.(((((....((.....))((((.((.....)))))).))))).)))))))....((((((....))))))...............--..-------------------- ( -32.20) >DroSec_CAF1 4043 102 - 1 UUACAGUUGGGAAUACCCCACCGAGAAGGGCCUACCGUUUGGCAGGCAGUGCUGCCAACUGAAAAUGGCUAUAAGAAGCCAAAAACAAUGUUCUUU--GU----------------GUUG ...(((((((.(.(((....((.....))((((.((....)).)))).))).).)))))))....(((((......)))))...((((......))--))----------------.... ( -31.10) >DroSim_CAF1 4015 102 - 1 UUACAGUUGGGAAUACCCCACCGAGAAGGGCCUACCGUUUGGCAGGCAGUGCUGCCAACUGAAAAUGGCUAUAAAUAGCCAAAAACAAUGUUCUUU--GU----------------GUUG ...(((((((.(.(((....((.....))((((.((....)).)))).))).).)))))))....((((((....))))))...((((......))--))----------------.... ( -32.00) >DroEre_CAF1 6399 94 - 1 UUACAGAUGGGAAAACCCCACCGAGAAGGGCCUACCGUGUGGGAUGUAGUGCUGCCAACUGGAAAUCGCUAGAAAUAGCCAAA-CUGAGGU-------------------------GUUG .......((((.....))))((.....))((((.(((..(((.(.(.....)).)))..))).....((((....))))....-...))))-------------------------.... ( -21.90) >DroYak_CAF1 4726 119 - 1 UCACAGUUUGGAAUACCCCACCGAGAAGGGCCUACCGUAUGGGAAGUAGUGCUGGCAAGGGAAAAUCGCUAGAAAUUGCCAAA-ACUACGUUCAUUCUGUAAUGGGUAUUUAUACUAUUU ....(((...((((((((.....((((((((...((.....))..(((((..((((((.((.......)).....))))))..-))))))))).)))).....))))))))..))).... ( -32.90) >consensus UUACAGUUGGGAAUACCCCACCGAGAAGGGCCUACCGUUUGGGAGGCAGUGCUGCCAACUGAAAAUGGCUAGAAAUAGCCAAAAACAAUGUUCUUU__GU________________GUUG ...(((((((.(.(((....((.....))((((.((....)).)))).))).).)))))))....((((((....))))))....................................... (-19.36 = -21.40 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:10 2006