
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,643,983 – 10,644,096 |
| Length | 113 |
| Max. P | 0.995113 |

| Location | 10,643,983 – 10,644,096 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.76 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10643983 113 + 22407834 A---AGGGAACGAAAUGGCGAUCGCAUUUAAUAUUGCGAUCUAUGCAAUAAUUUUAGUGGGUAGUUUGGUACCCAGCAAGACUCA--GGAAUGGGUUCCAUAUGCAUAUCCUGUAGAU .---.((((..........(((((((........)))))))((((((..........((((((......))))))....((((((--....)))))).....))))))))))...... ( -33.40) >DroSec_CAF1 24094 113 + 1 A---AGGGAACGAAAUGGCGAUCGCAUUUAAUAUUGCGAUCUGUUCAAUAUUUUUACUGGGUAUUGUGGUAGCCACCAAGACUCA--GAAAUGGGUUCCAUAUGCAGAUUCUCUAGAU .---(((((((....(((((((((((........)))))))....((((((((.....)))))))).....))))....((((((--....)))))).........).)))))).... ( -32.70) >DroSim_CAF1 24726 113 + 1 A---AGGGAACGAAAUGGCGAUCGCAUUUAAUAUUGCGAUCUGUUCAAUAUUUUUAAUGGGUAUUGUGGUAGCCACCAAGACUCG--GAAAUGGGUUCCAUAUGCAGAUACUCUAGAU .---..(((((....(((((((((((........)))))))....((((((((.....)))))))).....))))(((.......--....))))))))................... ( -29.40) >DroEre_CAF1 24406 91 + 1 A---AAGUAACGGAAUGGCGAUCGCAUUUAAAUUGGCGAUCUGUGCAAUCAUUUUACUCGGUAAUGUCUUACCCAGCAAGACUCAACGGCA---GUU--------------------- .---.(((((..((.((((((((((..........)))))).)).)).))...)))))((.....(((((.......)))))....))...---...--------------------- ( -17.40) >DroYak_CAF1 24496 110 + 1 AAACAAGAAGCGGAAUGGCGAUCGCAUUUAAUUUGGCGAUCUGUUCAAUAAUUUUACUCGGUACUGUGGUACCCAGUAAGCCUCA--GAUA---GUUCCUU---UACUUCCUAUAGUU ......((((.((((((((((((((..........))))))............(((((.(((((....))))).))))))))...--....---)))))..---..))))........ ( -29.21) >consensus A___AGGGAACGAAAUGGCGAUCGCAUUUAAUAUUGCGAUCUGUUCAAUAAUUUUACUGGGUAUUGUGGUACCCAGCAAGACUCA__GAAAUGGGUUCCAUAUGCAGAUCCUCUAGAU ......(((((........(((((((........)))))))................(((((((....)))))))...................)))))................... (-17.24 = -18.76 + 1.52)

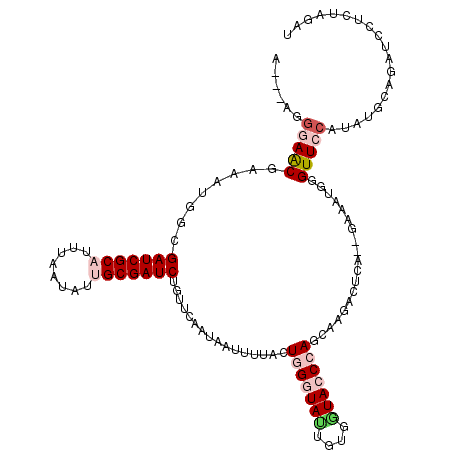

| Location | 10,643,983 – 10,644,096 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -18.38 |
| Energy contribution | -20.14 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10643983 113 - 22407834 AUCUACAGGAUAUGCAUAUGGAACCCAUUCC--UGAGUCUUGCUGGGUACCAAACUACCCACUAAAAUUAUUGCAUAGAUCGCAAUAUUAAAUGCGAUCGCCAUUUCGUUCCCU---U .......(((((((((...((((....))))--..(((.(((.((((((......)))))).))).)))..))))))(((((((........)))))))..........)))..---. ( -29.50) >DroSec_CAF1 24094 113 - 1 AUCUAGAGAAUCUGCAUAUGGAACCCAUUUC--UGAGUCUUGGUGGCUACCACAAUACCCAGUAAAAAUAUUGAACAGAUCGCAAUAUUAAAUGCGAUCGCCAUUUCGUUCCCU---U .....((((.((.....((((...))))...--.)).))))((((((............(((((....)))))....(((((((........))))))))))))).........---. ( -24.90) >DroSim_CAF1 24726 113 - 1 AUCUAGAGUAUCUGCAUAUGGAACCCAUUUC--CGAGUCUUGGUGGCUACCACAAUACCCAUUAAAAAUAUUGAACAGAUCGCAAUAUUAAAUGCGAUCGCCAUUUCGUUCCCU---U ...................((((((((....--.......)))((((.....(((((...........)))))....(((((((........)))))))))))....)))))..---. ( -23.80) >DroEre_CAF1 24406 91 - 1 ---------------------AAC---UGCCGUUGAGUCUUGCUGGGUAAGACAUUACCGAGUAAAAUGAUUGCACAGAUCGCCAAUUUAAAUGCGAUCGCCAUUCCGUUACUU---U ---------------------(((---(((.(((..((.(((((.(((((....))))).))))).))))).)))..((((((..........))))))........)))....---. ( -21.90) >DroYak_CAF1 24496 110 - 1 AACUAUAGGAAGUA---AAGGAAC---UAUC--UGAGGCUUACUGGGUACCACAGUACCGAGUAAAAUUAUUGAACAGAUCGCCAAAUUAAAUGCGAUCGCCAUUCCGCUUCUUGUUU ....(((((((((.---..(((((---....--.).((((((((.(((((....))))).)))))............((((((..........))))))))).))))))))))))).. ( -35.10) >consensus AUCUAGAGGAUCUGCAUAUGGAACCCAUUUC__UGAGUCUUGCUGGGUACCACAAUACCCAGUAAAAUUAUUGAACAGAUCGCAAUAUUAAAUGCGAUCGCCAUUUCGUUCCCU___U ...................(((((...............((((((((((......))))))))))............(((((((........)))))))........)))))...... (-18.38 = -20.14 + 1.76)

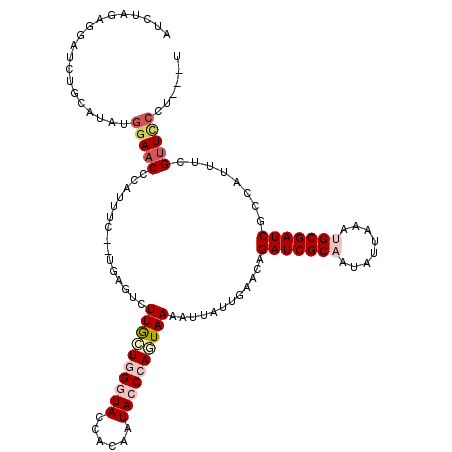
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:07 2006