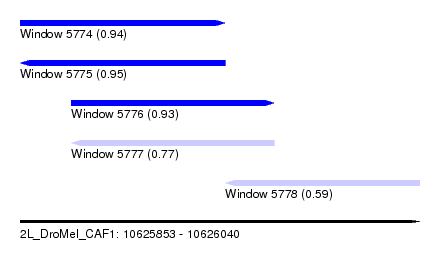
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,625,853 – 10,626,040 |
| Length | 187 |
| Max. P | 0.950566 |

| Location | 10,625,853 – 10,625,949 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942459 |
| Prediction | RNA |
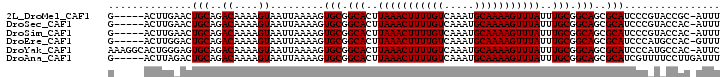
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10625853 96 + 22407834 G-----ACUUGAACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCGUACCGC-AUUU .-----((((.............))))......((((((((.((.(((((((((((.....)))))))))))..((((....))))....)).))))-)))) ( -25.72) >DroSec_CAF1 5886 96 + 1 G-----ACUUGAACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCGUACCAC-AUUU .-----...((..((((.(((((((((......(((((...)))))..)))))))))...((((((......))))))))))..))...........-.... ( -22.90) >DroSim_CAF1 5902 96 + 1 G-----ACUUGAACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCGUACCAC-AUUU .-----...((..((((.(((((((((......(((((...)))))..)))))))))...((((((......))))))))))..))...........-.... ( -22.90) >DroEre_CAF1 6374 96 + 1 G-----ACUUGGACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCAUGCCAC-GUUU .-----...(((..(((..((....)).........(((.(((..(((((((((((.....)))))))))))..))).)))..)))..)))......-.... ( -26.40) >DroYak_CAF1 6159 101 + 1 AAAGGCACUGGGAGUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCAUGCCAC-AUUC ...((((.(((((((((..((....)).........(((.(((..(((((((((((.....)))))))))))..))).))))))).)))))))))..-.... ( -41.30) >DroAna_CAF1 6184 97 + 1 G-----ACUUAGACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCGUUUUCCUUGAUUU .-----.....((.(((..((....)).........(((.(((..(((((((((((.....)))))))))))..))).)))..))))).............. ( -23.80) >consensus G_____ACUUGAACUGCAGACAAAAGUAAUUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUCCCGUACCAC_AUUU ..............(((..((....)).........(((.(((..(((((((((((.....)))))))))))..))).)))..)))................ (-22.42 = -22.42 + 0.00)
| Location | 10,625,853 – 10,625,949 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10625853 96 - 22407834 AAAU-GCGGUACGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUUCAAGU-----C ...(-((((((((.....)).))))))).....(((((((((...(((((((((..(((((...)))))......))))))))))))))...))))-----. ( -28.30) >DroSec_CAF1 5886 96 - 1 AAAU-GUGGUACGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUUCAAGU-----C ...(-((((((((.....)).))))))).....(((((((((...(((((((((..(((((...)))))......))))))))))))))...))))-----. ( -26.40) >DroSim_CAF1 5902 96 - 1 AAAU-GUGGUACGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUUCAAGU-----C ...(-((((((((.....)).))))))).....(((((((((...(((((((((..(((((...)))))......))))))))))))))...))))-----. ( -26.40) >DroEre_CAF1 6374 96 - 1 AAAC-GUGGCAUGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUCCAAGU-----C ....-..(((.(((..(((......(((((......)))))....(((((((((..(((((...)))))......))))))))).)))..))).))-----) ( -28.20) >DroYak_CAF1 6159 101 - 1 GAAU-GUGGCAUGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCACUCCCAGUGCCUUU ....-..((((((((((((......(((((......)))))....(((((((((..(((((...)))))......))))))))).))).))))).))))... ( -35.50) >DroAna_CAF1 6184 97 - 1 AAAUCAAGGAAAACGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUCUAAGU-----C ..............(((..(((((.(((((......)))))....(((((((((..(((((...)))))......))))))))).)))))....))-----) ( -24.30) >consensus AAAU_GUGGUACGGGAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAAUUACUUUUGUCUGCAGUCCAAGU_____C ................((.(((((.(((((......)))))....(((((((((..(((((...)))))......))))))))).))))).))......... (-23.32 = -23.52 + 0.19)


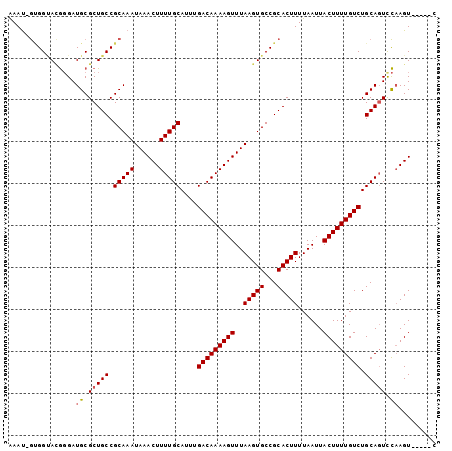
| Location | 10,625,877 – 10,625,972 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10625877 95 + 22407834 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC-CCGUACCGC-AUUU---UUCUAUCUUUUUAAGUCUUUGC-G ..((((((((((.((.(((((((((((.....)))))))))))..((((....))))..-..)).))))-))))---))....................-. ( -27.10) >DroPse_CAF1 6403 93 + 1 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCGAAAGUUUAUUUGCGGGAA----UCGGCAGGCAUC-AUUCUUUUCCCUUCUUCUCCAGC--UGGC-A .......(((.(((..(((((((((((.....)))))))))))..)))(((((----..((........-..))..)))))............--..))-) ( -22.50) >DroSec_CAF1 5910 95 + 1 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC-CCGUACCAC-AUUU---UUCUAUCUUUUUAAGUCUUUGC-C ((((((((((.(((..(((((((((((.....)))))))))))..))).)))(((....-.))).....-....---.......)))))))........-. ( -22.50) >DroSim_CAF1 5926 95 + 1 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC-CCGUACCAC-AUUU---UUCUAUCUUUUUAAGUCUUUGC-G ((((((((((.(((..(((((((((((.....)))))))))))..))).)))(((....-.))).....-....---.......)))))))........-. ( -22.50) >DroEre_CAF1 6398 98 + 1 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC-CCAUGCCAC-GUUUUCUUUUUAUAUUUUCCAGUCCUUGC-G ..(((((.((((((..(((((((((((.....)))))))))))..)))((((.......-...)))).)-))...)))))...................-. ( -25.00) >DroAna_CAF1 6208 94 + 1 UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC-GUUUUCCUUGAUUU---UUCUUUAUUUUU---UCUUUGUCG ...(((((((.(((..(((((((((((.....)))))))))))..))).)))....(((-(.......))))..---..))))......---......... ( -21.10) >consensus UUAAAAGUGCGGCACUUAAACUUUUGUCAAAUGCAAAAGUUUAUUUGCGGCAGCGCAUC_CCGUACCAC_AUUU___UUCUAUCUUUUUAAGUCUUUGC_G .......(((.(((..(((((((((((.....)))))))))))..))).)))................................................. (-19.60 = -19.63 + 0.03)

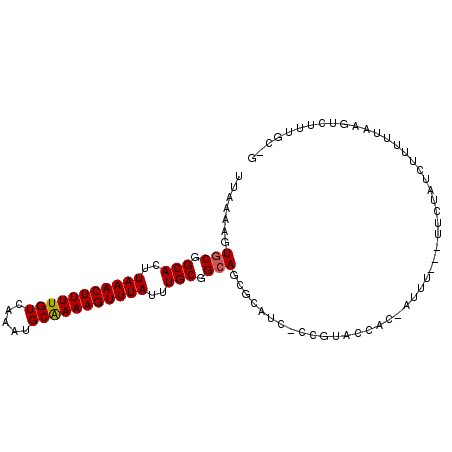
| Location | 10,625,877 – 10,625,972 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -16.50 |
| Energy contribution | -18.08 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10625877 95 - 22407834 C-GCAAAGACUUAAAAAGAUAGAA---AAAU-GCGGUACGG-GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA .-....................((---((.(-(((((((..-..((((....))))..(((((((((((....).)))))))))).)))))))).)))).. ( -28.00) >DroPse_CAF1 6403 93 - 1 U-GCCA--GCUGGAGAAGAAGGGAAAAGAAU-GAUGCCUGCCGA----UUCCCGCAAAUAAACUUUCGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA .-.((.--...))(((((..(((((..(...-.........)..----)))))(((..((((((((.((....).).))))))))..)))....))))).. ( -17.90) >DroSec_CAF1 5910 95 - 1 G-GCAAAGACUUAAAAAGAUAGAA---AAAU-GUGGUACGG-GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA .-....................((---((.(-(((((((..-..((((....))))..(((((((((((....).)))))))))).)))))))).)))).. ( -26.10) >DroSim_CAF1 5926 95 - 1 C-GCAAAGACUUAAAAAGAUAGAA---AAAU-GUGGUACGG-GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA .-....................((---((.(-(((((((..-..((((....))))..(((((((((((....).)))))))))).)))))))).)))).. ( -26.10) >DroEre_CAF1 6398 98 - 1 C-GCAAGGACUGGAAAAUAUAAAAAGAAAAC-GUGGCAUGG-GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA (-....)........................-(((((((..-..((((....))))..(((((((((((....).)))))))))).)))))))........ ( -25.10) >DroAna_CAF1 6208 94 - 1 CGACAAAGA---AAAAAUAAAGAA---AAAUCAAGGAAAAC-GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA ....((((.---........((..---..(((.........-)))...))((.(((..(((((((((((....).))))))))))..))).)).))))... ( -17.50) >consensus C_GCAAAGACUUAAAAAGAUAGAA___AAAU_GUGGUACGG_GAUGCGCUGCCGCAAAUAAACUUUUGCAUUUGACAAAAGUUUAAGUGCCGCACUUUUAA ................................(((((((.....((((....))))..(((((((((((....).)))))))))).)))))))........ (-16.50 = -18.08 + 1.58)

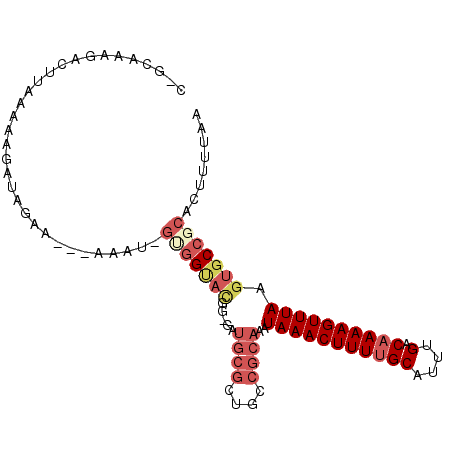
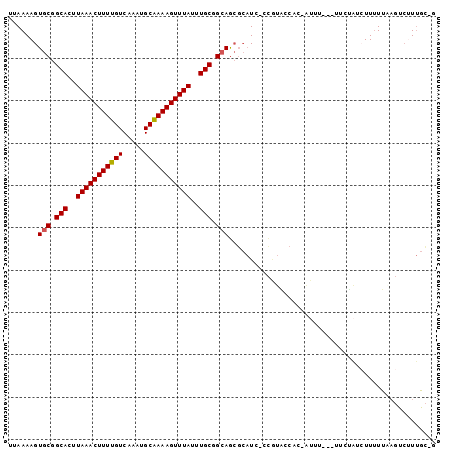
| Location | 10,625,949 – 10,626,040 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10625949 91 - 22407834 CACGUGUUAAAUGGCGCGUGAGCAGCUUGGGGAAUAAAUUGUGUAAGUGAAAAGGAUACACAGCAUCG------C-GCAAAGACUUAAAAAGAUAGAA--- ((((((((....))))))))......(((((........((((((...........))))))((....------.-)).....)))))..........--- ( -17.00) >DroPse_CAF1 6472 94 - 1 CACGUGUUAAAUGGCGCGUGAGCUGCCUGAAGAGUAAAUUCUGUAAGUGAAAGAGAUACACGACGCCUUC----U-GCCA--GCUGGAGAAGAAGGGAAAA ..(((((.....(((((....)).)))...........((((.........))))..)))))...(((((----(-.((.--...))...))))))..... ( -26.50) >DroSec_CAF1 5982 91 - 1 CACGUGUUAAAUGGCGCGUGAGCAGCUUGGGGAAUAAAUUGUGUAAGUGAAAAGGAUACACAGCAUCG------G-GCAAAGACUUAAAAAGAUAGAA--- ((((((((....))))))))....((((((........(((((((...........)))))))..)))------)-))....................--- ( -18.80) >DroEre_CAF1 6470 94 - 1 CACGUGUUAAAUGGCGCGUGAGCAGCUUGGGGAAUAAAUUGCGUAAGUGAAAAGGAUACACGGCACAG------C-GCAAGGACUGGAAAAUAUAAAAAGA ((((((((....))))))))..(((((...........((((((..(((....(......)..))).)------)-)))))).)))............... ( -19.60) >DroAna_CAF1 6281 95 - 1 CACGUGUUAAAUGGCGCGUGAGCAGCUUGGGGAAUAAAUUGUGUAAGUGAAAAAGAUACACAGCGGCGUCUUAGCGACAAAGA---AAAAAUAAAGAA--- ((((((((....)))))))).(..(((..((((.....(((((((..(....)...))))))).....)))))))..).....---............--- ( -21.00) >DroPer_CAF1 6647 94 - 1 CACGUGUUAAAUGGCGCGUGAGAUGCCUGAAGAGUAAAUUCUGUAAGUGAAAGAGAUACACGACGCCUUC----U-GCCA--GCUGGAGAAGAAGGGAAAA ..(((((.....(((((....).))))...........((((.........))))..)))))...(((((----(-.((.--...))...))))))..... ( -24.60) >consensus CACGUGUUAAAUGGCGCGUGAGCAGCUUGGGGAAUAAAUUGUGUAAGUGAAAAAGAUACACAGCACCG______C_GCAAAGACUGAAAAAGAAAGAA___ ((((((((....))))))))..................(((((((..(....)...)))))))...................................... (-12.66 = -12.47 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:04 2006