
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,621,032 – 10,621,131 |
| Length | 99 |
| Max. P | 0.753844 |

| Location | 10,621,032 – 10,621,131 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -18.59 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
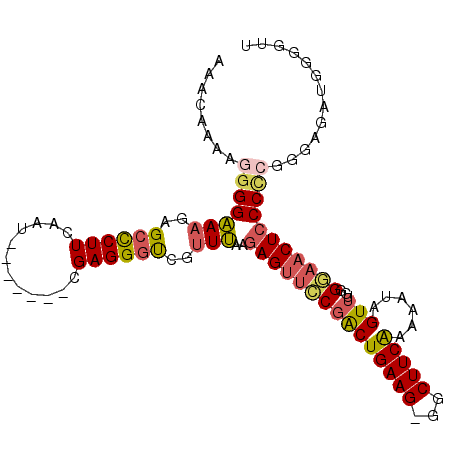
>2L_DroMel_CAF1 10621032 99 + 22407834 AAACAAAAGGGGAAAGAGCCCUUCAUU-------CGAGGGUCGUUUAAGAGUUCCGACUGAAG-GGCUUCAAAAAUAGUUUUGGGAACUCCCCCGGGAGUUGGGUUU ...(((((..(....((((((((((.(-------((..(.((......)).)..))).)))))-)))))......)..)))))..((((((....))))))...... ( -35.20) >DroSec_CAF1 1096 99 + 1 AAACAAAAGGGGAAAGAGCCCUUCAAU-------CGAGGGUCGGUUAAGAGUUCCGACUGAAG-GGCUUCAAAAAUAGUUUUGGGAACUCCCCCGGGAGUUGGGGUU ...(((((..(....((((((((((.(-------((..(.((......)).)..))).)))))-)))))......)..)))))..((((((....))))))...... ( -35.20) >DroSim_CAF1 1113 99 + 1 AAACAAAAGGGGAAAGAGCCCUUCAAU-------CGAGGCUCGUUUAAGAGUUCCGACUGAAG-GGCUUCAAAAAUAGUUUUGGGAACUCCCCCGGGAGUUGGGGUU ...(((((..(....((((((((((.(-------((..((((......))))..))).)))))-)))))......)..)))))..((((((....))))))...... ( -39.40) >DroEre_CAF1 1582 99 + 1 AAUAGAAAGGGGAAAGGGCCCUUCAAU-------CGAGGGUCGUUUAAAAGUUCCGGCUGAAG-GGCUUCAAAAAUAGUUCGGGAAACUCCCCAGGGAGAUGGGGUC ....(((........((((((((((..-------((.(((.(........))))))..)))))-))))).........)))((....))(((((......))))).. ( -31.63) >DroYak_CAF1 1236 99 + 1 AAACAAAAGGGGAAAGAGCCCUUCAAU-------CGAGGGUCGUUUAGGAGUUCCGGCUGAAG-GGCUUCAAAAAUAGUUUGGGGAACUCCCUCGGGAGAUGGGCUC ...............((((((.((..(-------(((((..(.....)(((((((((((....-))))(((((.....))))))))))))))))))..)).)))))) ( -35.90) >DroPer_CAF1 1056 104 + 1 AAACAAUGCCGGGAAGAGCUCUUCAGCUCUGCGACGAGGGCCGUUCAAGAGGGGCCACUGAAGGGGCUUCGAAAAUAGUUUGGGCAACUCCCA-GGAGGAAGGGA-- ......((((.(((((((((....))))))....(((((.((.((((.(.(....).))))).)).))))).......))).))))..((((.-.......))))-- ( -35.80) >consensus AAACAAAAGGGGAAAGAGCCCUUCAAU_______CGAGGGUCGUUUAAGAGUUCCGACUGAAG_GGCUUCAAAAAUAGUUUGGGGAACUCCCCCGGGAGAUGGGGUU ........(((((((..((((((............))))))..)))..(((((((((((((((...)))))......)))...)))))))))))............. (-18.59 = -19.02 + 0.42)

| Location | 10,621,032 – 10,621,131 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -14.78 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10621032 99 - 22407834 AAACCCAACUCCCGGGGGAGUUCCCAAAACUAUUUUUGAAGCC-CUUCAGUCGGAACUCUUAAACGACCCUCG-------AAUGAAGGGCUCUUUCCCCUUUUGUUU ......(((....(((((((....(((((....))))).((((-(((((.((((...((......))...)))-------).)))))))))..)))))))...))). ( -30.90) >DroSec_CAF1 1096 99 - 1 AACCCCAACUCCCGGGGGAGUUCCCAAAACUAUUUUUGAAGCC-CUUCAGUCGGAACUCUUAACCGACCCUCG-------AUUGAAGGGCUCUUUCCCCUUUUGUUU ......(((....(((((((....(((((....))))).((((-((((((((((...((......))...)))-------)))))))))))..)))))))...))). ( -34.60) >DroSim_CAF1 1113 99 - 1 AACCCCAACUCCCGGGGGAGUUCCCAAAACUAUUUUUGAAGCC-CUUCAGUCGGAACUCUUAAACGAGCCUCG-------AUUGAAGGGCUCUUUCCCCUUUUGUUU ......(((....(((((((....(((((....))))).((((-((((((((((..(((......)))..)))-------)))))))))))..)))))))...))). ( -38.40) >DroEre_CAF1 1582 99 - 1 GACCCCAUCUCCCUGGGGAGUUUCCCGAACUAUUUUUGAAGCC-CUUCAGCCGGAACUUUUAAACGACCCUCG-------AUUGAAGGGCCCUUUCCCCUUUCUAUU ..(((((......)))))(((((...))))).........(((-(((((..(((................)))-------..))))))))................. ( -24.89) >DroYak_CAF1 1236 99 - 1 GAGCCCAUCUCCCGAGGGAGUUCCCCAAACUAUUUUUGAAGCC-CUUCAGCCGGAACUCCUAAACGACCCUCG-------AUUGAAGGGCUCUUUCCCCUUUUGUUU ((((((.((...((((((((((((.((((.....))))..((.-.....)).)))))).........))))))-------...)).))))))............... ( -31.20) >DroPer_CAF1 1056 104 - 1 --UCCCUUCCUCC-UGGGAGUUGCCCAAACUAUUUUCGAAGCCCCUUCAGUGGCCCCUCUUGAACGGCCCUCGUCGCAGAGCUGAAGAGCUCUUCCCGGCAUUGUUU --..........(-((((((((.....))).......((.((..((((((.((((..........))))(((......))))))))).)))).))))))........ ( -26.30) >consensus AACCCCAACUCCCGGGGGAGUUCCCAAAACUAUUUUUGAAGCC_CUUCAGUCGGAACUCUUAAACGACCCUCG_______AUUGAAGGGCUCUUUCCCCUUUUGUUU .............(((((((((((...........((((((...))))))..)))))))......((((((((.........)).)))).))...))))........ (-14.78 = -15.42 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:59 2006