| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,620,433 – 10,620,541 |
| Length | 108 |
| Max. P | 0.898397 |

| Location | 10,620,433 – 10,620,541 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -24.24 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
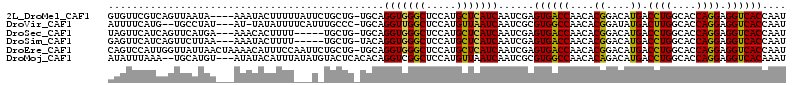
>2L_DroMel_CAF1 10620433 108 + 22407834 GUGUUCGUCAGUUAAUA----AAAUACUUUUUAUUCUGCUG-UGCAGGUGGGCUCCAUGCUCAUCAAUCGAGUGACCAACACGGACAUGACCUGGCACCAGGAGGUCACCAAU .(((....((((.((((----(((....)))))))..))))-.)))(((((((.....)))))))....(.((((((..((......)).((((....)))).)))))))... ( -34.00) >DroVir_CAF1 59648 106 + 1 AUUUUCAUG--UGCCUAU---AU-UAUAUUUUCAUUUGCCC-UGCAGGUUGGCUCCAUGUUAAUCAAUCGCGUGGCCAACACGGAUAUGACUUGGCACCAGGAGGUCACCAAU ((((((..(--((((...---.(-(((((((....((((..-.))))((((((..(((((.........)))))))))))..))))))))...)))))..))))))....... ( -27.00) >DroSec_CAF1 496 104 + 1 UAGUUCAUCAGUUCAUGA---AAACACUUUU-----UGCUG-UGCAGGUGGGCUCCAUGCUCAUCAAUCGAGUGACCAACACGGACAUGACCUGGCACCAGGAGGUCACCAAU ...(((((......))))---)..(((....-----....)-))..(((((((.....)))))))....(.((((((..((......)).((((....)))).)))))))... ( -29.40) >DroSim_CAF1 531 104 + 1 GAGUUCAUCAGUUCUUAA---AAAUACUUUU-----UGCUG-UACAGGUGGGCUCCAUGCUCAUCAAUCGAGUGACCAACACGGACAUGACCUGGCACCAGGAGGUCACCAAU ((((....((((....((---((....))))-----.))))-.)).(((((((.....)))))))..))(.((((((..((......)).((((....)))).)))))))... ( -28.70) >DroEre_CAF1 989 112 + 1 CAGUCCAUUGGUUAUUAACUAAAACAUUUCCAAUUCUGCUG-UGCAGGUGGGCUCCAUGCUCAUCAAUCGAGUGACCAACACGGACAUGACCUGGCACCAGGAGGUCACCAAU ..((((.(((((((((....................(((..-.)))(((((((.....))))))).....)))))))))...)))).((((((.........))))))..... ( -32.40) >DroMoj_CAF1 57293 108 + 1 AUAUUUAAA--UGCAUGU---AUAUACAUUUAUAUGUACUCACACAGGUCGGCUCCAUGUUAAUCAAUCGCGUGGCCAACACAGACAUGACCUGGCACCAGGAGGUCACAAAU .........--.((..((---(((((......))))))).......(((..((.....))..)))....))((((((....((....)).((((....)))).)))))).... ( -27.40) >consensus AAGUUCAUCAGUUCAUAA___AAAUACUUUU___U_UGCUG_UGCAGGUGGGCUCCAUGCUCAUCAAUCGAGUGACCAACACGGACAUGACCUGGCACCAGGAGGUCACCAAU ..............................................(((((((.....)))))))......((((((....((....)).((((....)))).)))))).... (-24.24 = -23.47 + -0.78)

| Location | 10,620,433 – 10,620,541 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.53 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
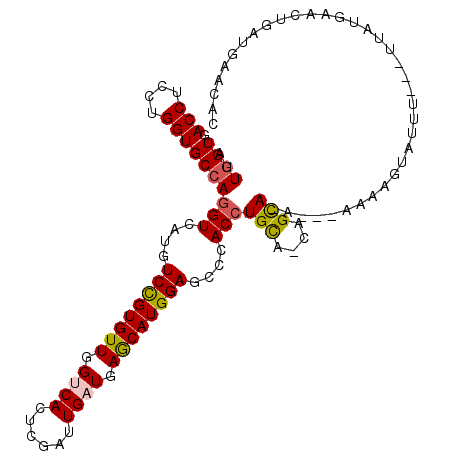
>2L_DroMel_CAF1 10620433 108 - 22407834 AUUGGUGACCUCCUGGUGCCAGGUCAUGUCCGUGUUGGUCACUCGAUUGAUGAGCAUGGAGCCCACCUGCA-CAGCAGAAUAAAAAGUAUUU----UAUUAACUGACGAACAC .(((((.(((....))))))))((((..((((((((.((((......)))).))))))))......((((.-..))))(((((((....)))----))))...))))...... ( -31.70) >DroVir_CAF1 59648 106 - 1 AUUGGUGACCUCCUGGUGCCAAGUCAUAUCCGUGUUGGCCACGCGAUUGAUUAACAUGGAGCCAACCUGCA-GGGCAAAUGAAAAUAUA-AU---AUAGGCA--CAUGAAAAU .(((((.(((....)))))))).((((.(((((((((..((......))..)))))))))(((..((....-))....((....))...-..---...))).--.)))).... ( -28.30) >DroSec_CAF1 496 104 - 1 AUUGGUGACCUCCUGGUGCCAGGUCAUGUCCGUGUUGGUCACUCGAUUGAUGAGCAUGGAGCCCACCUGCA-CAGCA-----AAAAGUGUUU---UCAUGAACUGAUGAACUA ...((((....((((....)))).....((((((((.((((......)))).))))))))...)))).(((-(....-----....)))).(---((((......)))))... ( -30.90) >DroSim_CAF1 531 104 - 1 AUUGGUGACCUCCUGGUGCCAGGUCAUGUCCGUGUUGGUCACUCGAUUGAUGAGCAUGGAGCCCACCUGUA-CAGCA-----AAAAGUAUUU---UUAAGAACUGAUGAACUC .(((.(((((....)))..(((((....((((((((.((((......)))).))))))))....)))))..-)).))-----).........---.................. ( -27.90) >DroEre_CAF1 989 112 - 1 AUUGGUGACCUCCUGGUGCCAGGUCAUGUCCGUGUUGGUCACUCGAUUGAUGAGCAUGGAGCCCACCUGCA-CAGCAGAAUUGGAAAUGUUUUAGUUAAUAACCAAUGGACUG ((((((......(((((((.((((....((((((((.((((......)))).))))))))....)))))))-)..)))(((((((.....)))))))....))))))...... ( -34.20) >DroMoj_CAF1 57293 108 - 1 AUUUGUGACCUCCUGGUGCCAGGUCAUGUCUGUGUUGGCCACGCGAUUGAUUAACAUGGAGCCGACCUGUGUGAGUACAUAUAAAUGUAUAU---ACAUGCA--UUUAAAUAU ...(((((((....)))((((((((...(((((((((..((......))..)))))))))...)))))).)).....))))(((((((((..---..)))))--))))..... ( -30.60) >consensus AUUGGUGACCUCCUGGUGCCAGGUCAUGUCCGUGUUGGUCACUCGAUUGAUGAGCAUGGAGCCCACCUGCA_CAGCA_A___AAAAGUAUUU___UUAUGAACUGAUGAACAC ..((((.(((....)))))))(((....((((((((.((((......)))).))))))))....)))(((....))).................................... (-21.59 = -21.53 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:57 2006