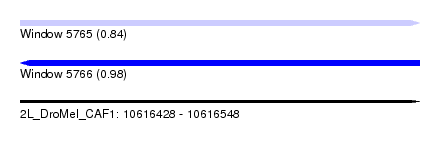
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,616,428 – 10,616,548 |
| Length | 120 |
| Max. P | 0.980570 |

| Location | 10,616,428 – 10,616,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
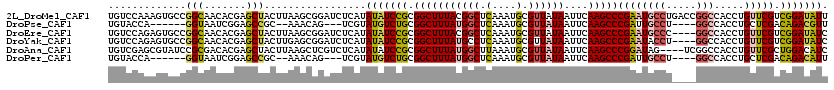
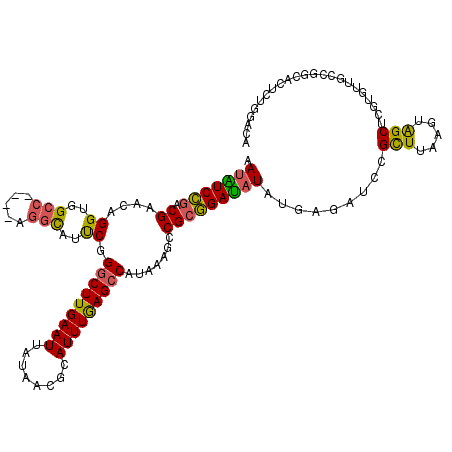
>2L_DroMel_CAF1 10616428 120 + 22407834 UGUCCAAAGUGCCGGCAACACGAGCUACUUAAGCGGAUCUCAUAUAUCCGCGGCUUUACGGCUCAAAUGCGUUAUAAUUCAAGCCCGAAUGCCUGACCGGCCACCUGUUCGUCGGAUAUU (((((.((((((((......)).)).))))..((((((.......))))))(((.....((((..(((........)))..)))).....))).(((..((.....))..)))))))).. ( -34.40) >DroPse_CAF1 96223 105 + 1 UGUACCA------GGUAAUCGGAGCCGC--AAACAG---UCGUAUGUCUGCGGCUUUAUGGCUCAAAUGCGUUAUAAUUCAAGCCCGAUUGCCU----GGCCACCUGCUCGACAGACGUU ....(((------(((((((((((((((--(.(((.---.....))).)))))))(((((((.(....).))))))).......))))))))))----))....(((.....)))..... ( -42.00) >DroEre_CAF1 96662 116 + 1 UGUCCAGAGUGCCGGCAACACGAGCUACUUAAGCGGAUCUCAUAUAUCCGCGGCUUUACGGCUCAAAUGCGUUAUAAUUCAAGCCCGAAUGCCC----GGCCACCUGUUCGUCGGAUAUC (((((........(....)((((((.......((((((.......))))))(((.....((((..(((........)))..))))((......)----))))....)))))).))))).. ( -33.30) >DroYak_CAF1 99507 116 + 1 UGUCCAGAGUGCCGGCAACACGAGCUACUUGAGCGGAUCUCAUAUAUCCGCGGCUUUAUGCCUCAAAUGCGUUAUAAUUCAAGCCCGAAUACCU----GGCCACCUGUUCGUCGGAUAUC ......((...(((((((((.(.((...((((((((((.......))))))(((.....)))))))..))............(((.(.....).----)))..).)))).)))))...)) ( -31.80) >DroAna_CAF1 97970 116 + 1 UGUCGAGCGUAUCCGCGACACGAGCUACUUAAGCUCGUCUCAUAUAUCCGCGGCUUUAUGGCUUAAAUGCGUUAUAAUUCAAGCCCGGAUAG----UCGGCCACCUGUUCGCUGGACAUC ((((.((((.((((((((.(((((((.....))))))).))...((((((.(((((((((((........))))))....))))))))))))----.)))......)).)))).)))).. ( -43.80) >DroPer_CAF1 98198 105 + 1 UGUACCA------GGUAAUCGGAGCCGC--AAACAG---UCGUAUGUCUGCGGCUUUAUGGCUCAAAUGCGUUAUAAUUCAAGCCCGAUUGCCU----GGCCACCUGCUCGACAGACAUU (((.(((------(((((((((((((((--(.(((.---.....))).)))))))(((((((.(....).))))))).......))))))))))----))....(((.....)))))).. ( -42.40) >consensus UGUCCAAAGUGCCGGCAACACGAGCUACUUAAGCGGAUCUCAUAUAUCCGCGGCUUUAUGGCUCAAAUGCGUUAUAAUUCAAGCCCGAAUGCCU____GGCCACCUGUUCGUCGGACAUC .............(((.......))).................(((((((.(((((((((((.(....).))))))....)))))((((((((.....))).....))))).))))))). (-18.18 = -18.30 + 0.12)
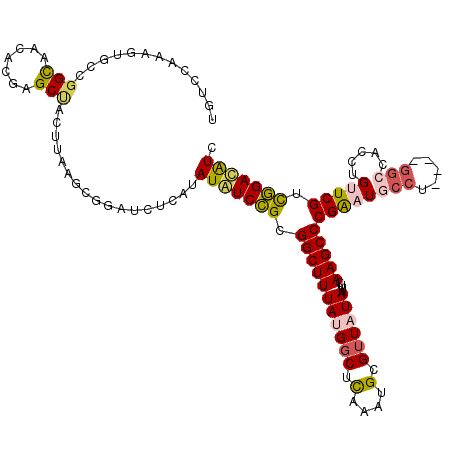
| Location | 10,616,428 – 10,616,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -21.74 |
| Energy contribution | -20.72 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10616428 120 - 22407834 AAUAUCCGACGAACAGGUGGCCGGUCAGGCAUUCGGGCUUGAAUUAUAACGCAUUUGAGCCGUAAAGCCGCGGAUAUAUGAGAUCCGCUUAAGUAGCUCGUGUUGCCGGCACUUUGGACA ....(((((......((((.(((((..(((.((..((((..(((........)))..))))..)).)))((((((.......))))))................)))))))))))))).. ( -39.30) >DroPse_CAF1 96223 105 - 1 AACGUCUGUCGAGCAGGUGGCC----AGGCAAUCGGGCUUGAAUUAUAACGCAUUUGAGCCAUAAAGCCGCAGACAUACGA---CUGUUU--GCGGCUCCGAUUACC------UGGUACA ..(..(((.....)))..)(((----(((.(((((((((..(((........)))..))))....(((((((((((.....---.)))))--)))))).))))).))------))))... ( -49.20) >DroEre_CAF1 96662 116 - 1 GAUAUCCGACGAACAGGUGGCC----GGGCAUUCGGGCUUGAAUUAUAACGCAUUUGAGCCGUAAAGCCGCGGAUAUAUGAGAUCCGCUUAAGUAGCUCGUGUUGCCGGCACUCUGGACA ....((((.......((..(((----((((.....((((..(((........)))..))))........((((((.......)))))).......))))).))..)).......)))).. ( -39.54) >DroYak_CAF1 99507 116 - 1 GAUAUCCGACGAACAGGUGGCC----AGGUAUUCGGGCUUGAAUUAUAACGCAUUUGAGGCAUAAAGCCGCGGAUAUAUGAGAUCCGCUCAAGUAGCUCGUGUUGCCGGCACUCUGGACA ..........(..((((((.((----.((((..((((((.............(((((((((.....)))((((((.......))))))))))))))))))...))))))))).)))..). ( -39.01) >DroAna_CAF1 97970 116 - 1 GAUGUCCAGCGAACAGGUGGCCGA----CUAUCCGGGCUUGAAUUAUAACGCAUUUAAGCCAUAAAGCCGCGGAUAUAUGAGACGAGCUUAAGUAGCUCGUGUCGCGGAUACGCUCGACA ..((((.((((.....(((((((.----.....))(((((((((........))))))))).....)))))........((.(((((((.....))))))).)).......)))).)))) ( -43.60) >DroPer_CAF1 98198 105 - 1 AAUGUCUGUCGAGCAGGUGGCC----AGGCAAUCGGGCUUGAAUUAUAACGCAUUUGAGCCAUAAAGCCGCAGACAUACGA---CUGUUU--GCGGCUCCGAUUACC------UGGUACA ..(..(((.....)))..)(((----(((.(((((((((..(((........)))..))))....(((((((((((.....---.)))))--)))))).))))).))------))))... ( -47.80) >consensus AAUAUCCGACGAACAGGUGGCC____AGGCAUUCGGGCUUGAAUUAUAACGCAUUUGAGCCAUAAAGCCGCGGAUAUAUGAGAUCCGCUUAAGUAGCUCGUGUUGCCGGCACUCUGGACA .(((((((.((....((..(((.....)))..)).(((((((((........))))))))).......))))))))).........(((.....)))....................... (-21.74 = -20.72 + -1.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:53 2006