| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,616,292 – 10,616,402 |
| Length | 110 |
| Max. P | 0.727480 |

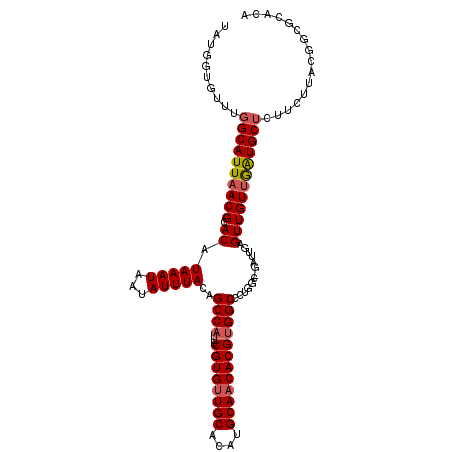
| Location | 10,616,292 – 10,616,402 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.72 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -25.91 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10616292 110 + 22407834 UGUGCGCCGUAAGAAGAGCAUCAACAACUCAAUC-GCCAAAGCCACGUGUUGCAUGUGCAACACGAAAUGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGCCAAACACCAUA ...((((((((((..(((.........)))....-.....(((((((((((((....))))))))...)))))..........)))))).))))................. ( -29.50) >DroSec_CAF1 95845 103 + 1 UGUGCGCCGUAAGAAGAGCAUCAACAACUCAAUC-GCCAGGGCCACGUGUUGCAUGUGCAACACGAAAUGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGC-------CAUA ...((((((((((..(((.........)))....-...(.(((((((((((((....))))))))...))))).)........)))))).))))......-------.... ( -29.90) >DroSim_CAF1 96325 110 + 1 UGUGCGCCGUAAGAAGAGCAUCAACAACUCAAUU-GCCAGGGCCACGUGUUGCAUGUGCAACACGAAAUGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGCCAAACACCAUA ...((((((((((..(((.........)))..((-((...(((((((((((((....))))))))...)))))))))......)))))).))))................. ( -30.30) >DroEre_CAF1 96521 110 + 1 UGUGGGCCGUAAGAAGAGCAUUAACAACUCAAUC-GCCAGGGCCACGUGUUGCAUGUGCAACACGAAAAGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGCCAAACACCAUA (((((..(....)..(.((((((((.........-.....((((.((((((((....))))))))....))))(((.(((.....))).)))))))))))).....))))) ( -34.20) >DroYak_CAF1 99366 110 + 1 UGUGCGCCGUAAGCAGAGCAUCAACAACUCAAUC-GCCAGGGCCACGUGUUGCAUGUGCAGCACGAAAUGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGCCAAACACCGUA ..(((.......)))(.((((.(((.........-.....(((((((((((((....))))))))...)))))(((.(((.....))).)))))).))))).......... ( -29.80) >DroAna_CAF1 97826 111 + 1 GCUGGUCUGCAAGACGAGCACCUACAACUCAAUGGGCCAGGGCCACGUGUUGCAUGUGCAACACGAAAUGGCUGUAAAUAAUAUUUAUGUGCGUUAAUGCCAAACACCAUA (((.(((.....))).)))............((((...(.(((((((((((((....))))))))...))))).)............((.(((....)))))....)))). ( -34.40) >consensus UGUGCGCCGUAAGAAGAGCAUCAACAACUCAAUC_GCCAGGGCCACGUGUUGCAUGUGCAACACGAAAUGGCUGUAAAUAUUAUUUAUGUGCGUUAAUGCCAAACACCAUA .................((((.(((...............(((((((((((((....))))))))...)))))(((.(((.....))).)))))).))))........... (-25.91 = -26.13 + 0.22)

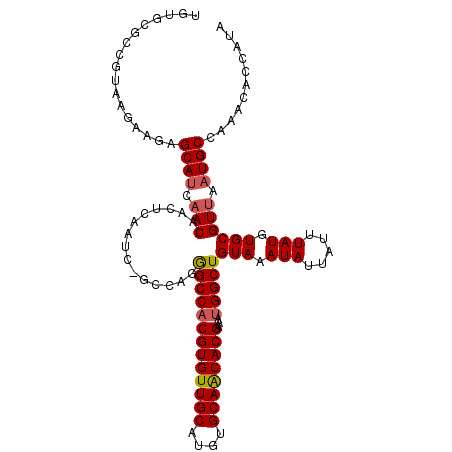

| Location | 10,616,292 – 10,616,402 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.72 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10616292 110 - 22407834 UAUGGUGUUUGGCAUUAACGCACAUAAAUAAUAUUUACAGCCAUUUCGUGUUGCACAUGCAACACGUGGCUUUGGC-GAUUGAGUUGUUGAUGCUCUUCUUACGGCGCACA (((((((((.......))))).)))).............(((....((((((((....)))))))).(((.(..((-(((...)))))..).)))........)))..... ( -31.90) >DroSec_CAF1 95845 103 - 1 UAUG-------GCAUUAACGCACAUAAAUAAUAUUUACAGCCAUUUCGUGUUGCACAUGCAACACGUGGCCCUGGC-GAUUGAGUUGUUGAUGCUCUUCUUACGGCGCACA ...(-------(((((((((.((.(((((...)))))..((((...((((((((....))))))))......))))-......))))))))))))................ ( -28.90) >DroSim_CAF1 96325 110 - 1 UAUGGUGUUUGGCAUUAACGCACAUAAAUAAUAUUUACAGCCAUUUCGUGUUGCACAUGCAACACGUGGCCCUGGC-AAUUGAGUUGUUGAUGCUCUUCUUACGGCGCACA ....(((((.((((((((((.((.(((((...)))))..((((...((((((((....))))))))......))))-......))))))))))))........)))))... ( -30.90) >DroEre_CAF1 96521 110 - 1 UAUGGUGUUUGGCAUUAACGCACAUAAAUAAUAUUUACAGCCUUUUCGUGUUGCACAUGCAACACGUGGCCCUGGC-GAUUGAGUUGUUAAUGCUCUUCUUACGGCCCACA (((((((((.......))))).)))).....................(((((((....)))))))((((((.(((.-((..((((.......)))).))))).)).)))). ( -30.80) >DroYak_CAF1 99366 110 - 1 UACGGUGUUUGGCAUUAACGCACAUAAAUAAUAUUUACAGCCAUUUCGUGCUGCACAUGCAACACGUGGCCCUGGC-GAUUGAGUUGUUGAUGCUCUGCUUACGGCGCACA ....((((..((((((((((.((.(((((...)))))..((((...((((.(((....))).))))......))))-......))))))))))))..((.....)))))). ( -29.80) >DroAna_CAF1 97826 111 - 1 UAUGGUGUUUGGCAUUAACGCACAUAAAUAUUAUUUACAGCCAUUUCGUGUUGCACAUGCAACACGUGGCCCUGGCCCAUUGAGUUGUAGGUGCUCGUCUUGCAGACCAGC (((((((((.......))))).))))......(((((((((((...((((((((....)))))))).(((....)))...)).)))))))))(((.(((.....))).))) ( -37.20) >consensus UAUGGUGUUUGGCAUUAACGCACAUAAAUAAUAUUUACAGCCAUUUCGUGUUGCACAUGCAACACGUGGCCCUGGC_GAUUGAGUUGUUGAUGCUCUUCUUACGGCGCACA ..........((((((((((.((.(((((...)))))..((((...((((((((....)))))))))))).............))))))))))))................ (-26.19 = -26.58 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:51 2006