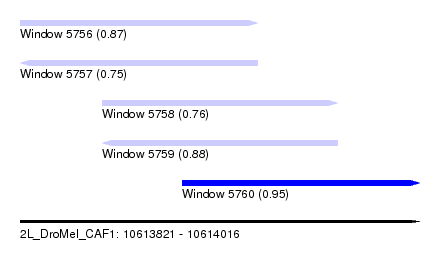
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,613,821 – 10,614,016 |
| Length | 195 |
| Max. P | 0.946220 |

| Location | 10,613,821 – 10,613,937 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -19.85 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10613821 116 + 22407834 UCACCCCUCUAAGUAAUACUUCCUUAUGAUAUACAAGUUCCCCCAUUUUCGUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAAC ...................((((((((((...................))))))).))).((((((......((((((.....)))))).....(((((.....))))))))))). ( -26.71) >DroSec_CAF1 93441 104 + 1 UCACCCCUCUA-GUAAAACUUCUU-----------ACUUCCCCCAUUUUCGUGAGCGAAUUUGCGCACUUUAGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAAC ..........(-((((......))-----------))).........((((....)))).((((((...(((((((((.....)))))))))..(((((.....))))))))))). ( -27.60) >DroSim_CAF1 93823 115 + 1 UCACCCCUCUA-GUAAUACAUCCUUUUGAUAUACAAGUUCCCCCAUUUUUUUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAAC ...........-(((...((......))...)))((((((.(.((......)).).))))))((((......((((((.....)))))).....(((((.....)))))))))... ( -22.90) >DroEre_CAF1 94112 114 + 1 UCACCCCUC-A-AUAAUACUUCUUUUUGAUAUACGAGUUCCCCCAUUUUGGUGAGCGAUUUUGCGCAUUUUUAUGACACUAAAUGUCACUAAUUUGACAAGAAAUGUCAGCGCAAC .........-.-........................((((.((......)).))))....((((((.......(((((.....)))))......(((((.....))))))))))). ( -22.90) >DroYak_CAF1 96811 115 + 1 UCACCCCUCUA-AUAAUACUGCUUUUUGAUAUACAAGCUCCCCCAUUUUGGUGAGCGAAUUUGCGCACUUUUAUGACACUAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAAC ...........-........................((((.((......)).))))....((((((.......(((((.....)))))......(((((.....))))))))))). ( -25.10) >DroAna_CAF1 95501 115 + 1 UCACCCCUCUA-AUAAUACUUUCUUUUGAAAUACAAGCUCCCCCAUUCGCCUGGCCAAAUUUGCGCAUUUUCGUGACACUAAAUGUCACUAAUUUGACAGAAAAUGUCAGCGCAGA ...........-.......((((....))))..((.((..........)).))......(((((((......((((((.....)))))).....(((((.....)))))))))))) ( -24.00) >consensus UCACCCCUCUA_AUAAUACUUCCUUUUGAUAUACAAGUUCCCCCAUUUUCGUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAAC ....................................((((.(........).))))....((((((......((((((.....)))))).....(((((.....))))))))))). (-19.85 = -20.47 + 0.61)

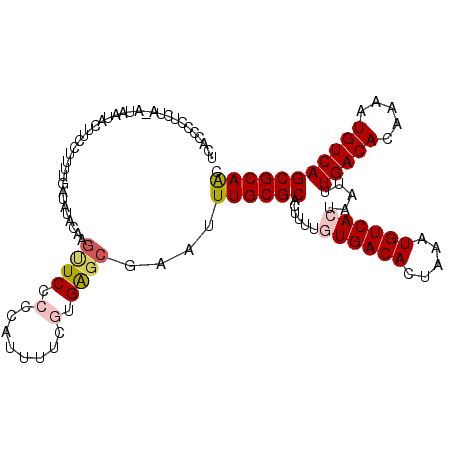
| Location | 10,613,821 – 10,613,937 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -25.43 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10613821 116 - 22407834 GUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCACGAAAAUGGGGGAACUUGUAUAUCAUAAGGAAGUAUUACUUAGAGGGGUGA .((((((((((((...)))))).....(((((((...)))))))......))))))..((((((......(..((....))..)...((.((((.........)))))).)))))) ( -33.50) >DroSec_CAF1 93441 104 - 1 GUUGCGCUGACAUUUUGUGUCAAAUUAGUGACAUUUAGUGUCACUAAAGUGCGCAAAUUCGCUCACGAAAAUGGGGGAAGU-----------AAGAAGUUUUAC-UAGAGGGGUGA .((((((((((((...))))))..((((((((((...))))))))))...))))))..((((((.(.....(((..(((.(-----------....).)))..)-))..))))))) ( -33.20) >DroSim_CAF1 93823 115 - 1 GUUGCGCUGACAUUUUGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCAAAAAAAUGGGGGAACUUGUAUAUCAAAAGGAUGUAUUAC-UAGAGGGGUGA .(((((((((((((..((((((......))))))..))))))).......))))))..((((((........(((....)))(((((((.....)))))))...-.....)))))) ( -34.61) >DroEre_CAF1 94112 114 - 1 GUUGCGCUGACAUUUCUUGUCAAAUUAGUGACAUUUAGUGUCAUAAAAAUGCGCAAAAUCGCUCACCAAAAUGGGGGAACUCGUAUAUCAAAAAGAAGUAUUAU-U-GAGGGGUGA .(((((((((((((...(((((......)))))...))))))).......))))))..((((((..(((...(((....)))((((.((.....)).))))..)-)-)..)))))) ( -34.01) >DroYak_CAF1 96811 115 - 1 GUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCAUAAAAGUGCGCAAAUUCGCUCACCAAAAUGGGGGAGCUUGUAUAUCAAAAAGCAGUAUUAU-UAGAGGGGUGA .(((((((((((((..((((((......))))))..))))))).......))))))..((((((.((.....))....((((..........))))........-.....)))))) ( -32.81) >DroAna_CAF1 95501 115 - 1 UCUGCGCUGACAUUUUCUGUCAAAUUAGUGACAUUUAGUGUCACGAAAAUGCGCAAAUUUGGCCAGGCGAAUGGGGGAGCUUGUAUUUCAAAAGAAAGUAUUAU-UAGAGGGGUGA ..((((((((((.....))))).....(((((((...)))))))......)))))......(((...(..(..((....))..).((((....)))).......-..)...))).. ( -27.30) >consensus GUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCACGAAAAUGGGGGAACUUGUAUAUCAAAAAGAAGUAUUAC_UAGAGGGGUGA .(((((((((((.....))))).....(((((((...)))))))......)))))).(((.((((......)))).)))..................................... (-25.43 = -25.27 + -0.17)
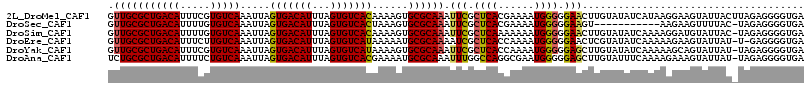
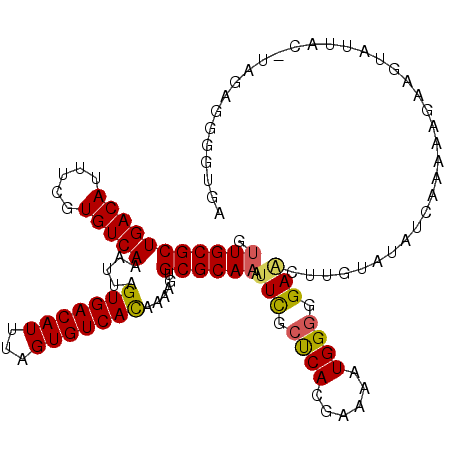
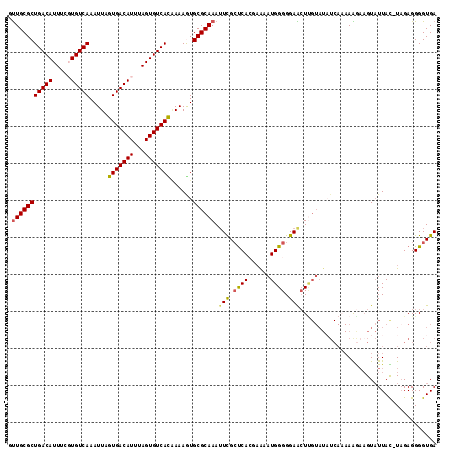
| Location | 10,613,861 – 10,613,976 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10613861 115 + 22407834 CCCCAUUUUCGUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUG ..((((.(((((.((.....((((((......((((((.....)))))).....(((((.....)))))))))))))...))))).))))..((((.((..-....)))))).... ( -31.70) >DroSec_CAF1 93469 115 + 1 CCCCAUUUUCGUGAGCGAAUUUGCGCACUUUAGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUG ..((((.(((((.((.....((((((...(((((((((.....)))))))))..(((((.....)))))))))))))...))))).))))..((((.((..-....)))))).... ( -35.10) >DroSim_CAF1 93862 115 + 1 CCCCAUUUUUUUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUG ...((((((((((((((...((((((......((((((.....)))))).....(((((.....)))))))))))....)))...(((.....))))))))-))))))........ ( -30.30) >DroEre_CAF1 94150 115 + 1 CCCCAUUUUGGUGAGCGAUUUUGCGCAUUUUUAUGACACUAAAUGUCACUAAUUUGACAAGAAAUGUCAGCGCAACUAAUGCGAGUAUGGAAUGUGUCAGA-AAAAUGCAUAACUG .........(((..(((....)))((((((((.((((((....(((.(((...((((((.....))))))((((.....))))))))))....)))))).)-)))))))...))). ( -29.40) >DroYak_CAF1 96850 115 + 1 CCCCAUUUUGGUGAGCGAAUUUGCGCACUUUUAUGACACUAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUG ...((((((((((.(((....))).))))....(((((.....)))))....(((((((((..((((...((((.....)))).))))....)))))))))-))))))........ ( -26.90) >DroAna_CAF1 95540 116 + 1 CCCCAUUCGCCUGGCCAAAUUUGCGCAUUUUCGUGACACUAAAUGUCACUAAUUUGACAGAAAAUGUCAGCGCAGAUAAUGCGAAUAUGGAAUGUGUCGGAUAAAAUGCAUAACUG ((..((((((........((((((((......((((((.....)))))).....(((((.....)))))))))))))...))))))..)).((((((........))))))..... ( -32.00) >consensus CCCCAUUUUCGUGAGCGAAUUUGCGCACUUUUGUGACACUAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA_AAAAUGCAUAACUG ..((((........(((...((((((......((((((.....)))))).....(((((.....)))))))))))....)))....))))..((((.((.......)))))).... (-24.92 = -25.00 + 0.08)

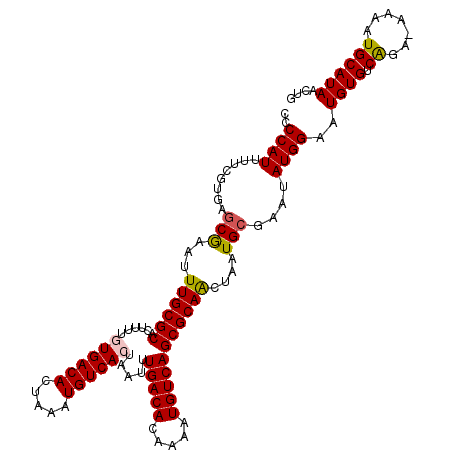
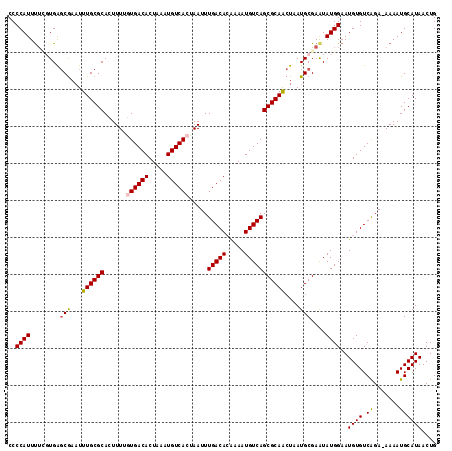
| Location | 10,613,861 – 10,613,976 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -25.73 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10613861 115 - 22407834 CAGUUAUGCAUUUU-UCUGACACAUUCCAUAUUCGCAUUAGUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCACGAAAAUGGGG (((..(......).-.)))......(((((.((((((.....)))(.(((((((..((((((......))))))..))))))))....(((.((......)).)))))).))))). ( -30.90) >DroSec_CAF1 93469 115 - 1 CAGUUAUGCAUUUU-UCUGACACAUUCCAUAUUCGCAUUAGUUGCGCUGACAUUUUGUGUCAAAUUAGUGACAUUUAGUGUCACUAAAGUGCGCAAAUUCGCUCACGAAAAUGGGG ......(.((((((-(.(((((((.........((((.....)))).........)))))))..((((((((((...)))))))))).(((.((......)).)))))))))).). ( -34.27) >DroSim_CAF1 93862 115 - 1 CAGUUAUGCAUUUU-UCUGACACAUUCCAUAUUCGCAUUAGUUGCGCUGACAUUUUGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCAAAAAAAUGGGG .......(((((((-(.(((((((.........((((.....)))).........))))))).....(((((((...))))))))))))))).........((((......)))). ( -31.17) >DroEre_CAF1 94150 115 - 1 CAGUUAUGCAUUUU-UCUGACACAUUCCAUACUCGCAUUAGUUGCGCUGACAUUUCUUGUCAAAUUAGUGACAUUUAGUGUCAUAAAAAUGCGCAAAAUCGCUCACCAAAAUGGGG .......(((((((-(.((((((..........((((.....)))).(((((.....)))))...............)))))).))))))))((......))...((.....)).. ( -29.40) >DroYak_CAF1 96850 115 - 1 CAGUUAUGCAUUUU-UCUGACACAUUCCAUAUUCGCAUUAGUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCAUAAAAGUGCGCAAAUUCGCUCACCAAAAUGGGG .......(((((((-(.((((((..........((((.....))))..........((((((......))))))...)))))).))))))))((......))...((.....)).. ( -29.70) >DroAna_CAF1 95540 116 - 1 CAGUUAUGCAUUUUAUCCGACACAUUCCAUAUUCGCAUUAUCUGCGCUGACAUUUUCUGUCAAAUUAGUGACAUUUAGUGUCACGAAAAUGCGCAAAUUUGGCCAGGCGAAUGGGG ..........................((.(((((((...((.((((((((((.....))))).....(((((((...)))))))......))))).))........))))))))). ( -29.50) >consensus CAGUUAUGCAUUUU_UCUGACACAUUCCAUAUUCGCAUUAGUUGCGCUGACAUUUCGUGUCAAAUUAGUGACAUUUAGUGUCACAAAAGUGCGCAAAUUCGCUCACGAAAAUGGGG .......(((((((...((((((..........((((.....)))).(((((.....)))))...............))))))..))))))).........((((......)))). (-25.73 = -25.70 + -0.03)
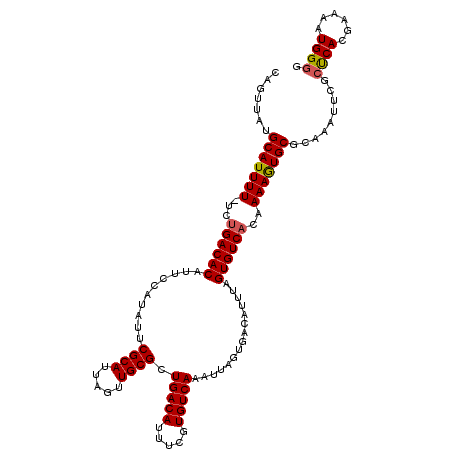
| Location | 10,613,900 – 10,614,016 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10613900 116 + 22407834 UAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUGGCGAGCUUUUUACCAGUUUUCUGUCGCAUUAGUGGCCACA .....((((((((.((((((.....))))))((((.....)))).........((((.((((-((........(((((..((...))..))))))))))).)))))))))))).... ( -31.60) >DroSec_CAF1 93508 116 + 1 UAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUGGCGAGCUUUUUACCAGUAUUCUGUCGCAUUAGUGGCCACA .....(((((((((((((((((..((((...((((.....)))).))))....)))))))))-....(((((((((((..((...))..)))))....))).))))))))))).... ( -32.30) >DroSim_CAF1 93901 116 + 1 UAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUGGCGAGCUUUUUACCAGUUUUCUGUCGCAUUAGUGGCCACA .....(((((((((((((((((..((((...((((.....)))).))))....)))))))))-....(((((((((((..((...))..)))))....))).))))))))))).... ( -32.30) >DroEre_CAF1 94189 116 + 1 UAAAUGUCACUAAUUUGACAAGAAAUGUCAGCGCAACUAAUGCGAGUAUGGAAUGUGUCAGA-AAAAUGCAUAACUGGCAAGCUUUCUACCAGCUUUCUGUCGCAUUAGUGGCCACA .....(((((((((.(((((.((((((((((((((.....)))).........((((.((..-....)))))).)))))).(((.......)))))))))))).))))))))).... ( -33.70) >DroYak_CAF1 96889 116 + 1 UAAAUGUCACUAAUUUGACACGAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA-AAAAUGCAUAACUGGCAAGUUUUUUACCAGCUUUCUGUCGCAUUAGUGGACACA ....((((......((((((.....))))))....((((((((((....((((.((((.(((-((..(((.......)))..))))).))..)).)))).)))))))))).)))).. ( -30.20) >DroAna_CAF1 95579 117 + 1 UAAAUGUCACUAAUUUGACAGAAAAUGUCAGCGCAGAUAAUGCGAAUAUGGAAUGUGUCGGAUAAAAUGCAUAACUGGCUAGCUUCUCCCCAACAAUCUGUCGCAUUAGAUGCCACA ..............((((((.....)))))).(((..((((((((....(((.(((...(((......((.......)).......)))...))).))).))))))))..))).... ( -28.22) >consensus UAAAUGUCACUAAUUUGACACAAAAUGUCAGCGCAACUAAUGCGAAUAUGGAAUGUGUCAGA_AAAAUGCAUAACUGGCGAGCUUUUUACCAGCUUUCUGUCGCAUUAGUGGCCACA .....(((((((((.(((((......(((((((((.....)))).........((((.((.......)))))).)))))(((((.......)))))..))))).))))))))).... (-26.36 = -26.42 + 0.06)

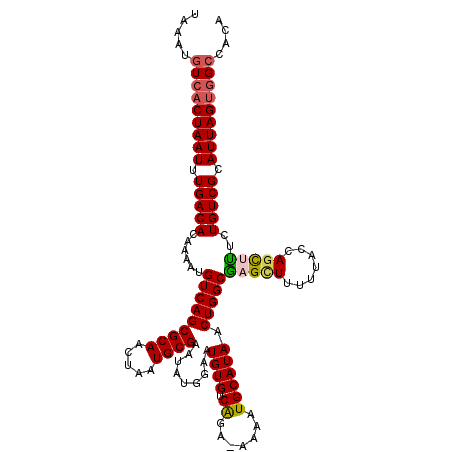
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:47 2006