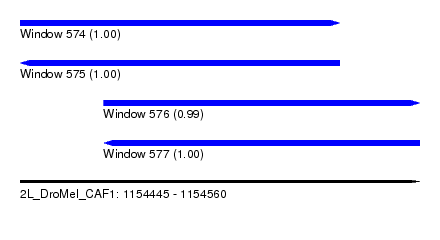
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,154,445 – 1,154,560 |
| Length | 115 |
| Max. P | 0.999555 |

| Location | 1,154,445 – 1,154,537 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.11 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
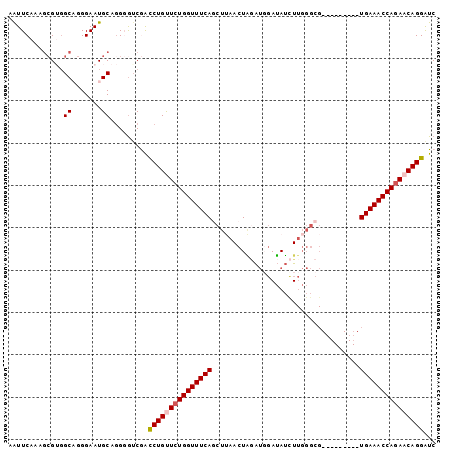
>2L_DroMel_CAF1 1154445 92 + 22407834 AAUUCAAAGCGGGGCUGGGAAUGCAGGGGUCGACCUGUUCUGGUUUCAGCUUAACUAUAUGGAUAUUUUGGGCG---------UGAAACCAAAACAGGAUC .((((..(((...)))..))))...........((((((.((((((((((((((..(((....))).)))))).---------)))))))).))))))... ( -29.60) >DroSec_CAF1 47961 92 + 1 AAUUCAAAGCGUUGCAGGGAAUGCAGGGGACGACCUGUUCUGGUUUCAGCUUAACUAGAUGGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUC .........(.(((((.....))))).).....((((((((((((((((((((...((((....))))))))).---------)))))))))))))))... ( -38.10) >DroSim_CAF1 43005 92 + 1 AAUUCAAAGCGGGGCUGGGAAUGCAAGGAUCGAUCUGUUCUGGUUUCAGCUUAACUAGAUUGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUC .((((..(((...)))..)))).....((((...(((((((((((((((((((...((((....))))))))).---------)))))))))))))))))) ( -35.00) >DroEre_CAF1 41643 100 + 1 AGUUCAAAGCGUGGCAGGGAAUGCAGG-GUCAGCCUGCUCUGGUUUCAACUAAACUGGAUGGAUAUCUUGGGCGCAGUUGCAGUGAAACCAGAACAGGGUC ........((((........))))...-.....((((.((((((((((....((((((..((....))....).)))))....)))))))))).))))... ( -32.20) >DroYak_CAF1 42767 85 + 1 AGUUCAAGCAGUGGCAGGGAAUGCAGG-GUCAGCCUGCUCUGGUUUCAACUAAACUUAGACCAGAACU---------------UGAAACCAGAACAGGAUC .((((.....((..((((....(((((-.....)))))((((((((...........)))))))).))---------------))..))..))))...... ( -23.90) >consensus AAUUCAAAGCGUGGCAGGGAAUGCAGGGGUCGACCUGUUCUGGUUUCAGCUUAACUAGAUGGAUAUCUUGGGCG_________UGAAACCAGAACAGGAUC .............((.......)).........(((((((((((((((...................................)))))))))))))))... (-20.67 = -21.11 + 0.44)

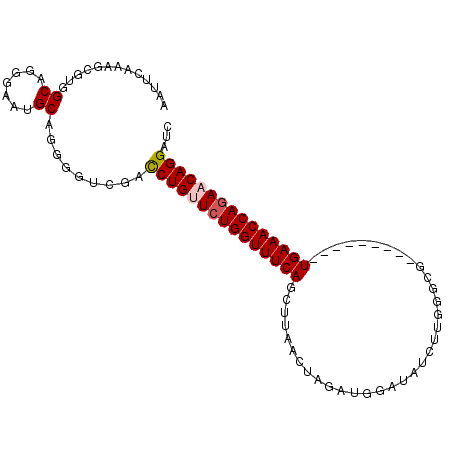
| Location | 1,154,445 – 1,154,537 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.59 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1154445 92 - 22407834 GAUCCUGUUUUGGUUUCA---------CGCCCAAAAUAUCCAUAUAGUUAAGCUGAAACCAGAACAGGUCGACCCCUGCAUUCCCAGCCCCGCUUUGAAUU ((.(((((((((((((((---------.((......(((....))).....))))))))))))))))))).........((((..(((...)))..)))). ( -26.70) >DroSec_CAF1 47961 92 - 1 GAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGUCCCCUGCAUUCCCUGCAACGCUUUGAAUU ((.(((((((((((((((---------.((.(.((((....)))).)....)))))))))))))))))))......((((.....))))............ ( -31.80) >DroSim_CAF1 43005 92 - 1 GAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCAAUCUAGUUAAGCUGAAACCAGAACAGAUCGAUCCUUGCAUUCCCAGCCCCGCUUUGAAUU ((((.(((((((((((((---------.((.(.((((....)))).)....))))))))))))))))))).........((((..(((...)))..)))). ( -29.30) >DroEre_CAF1 41643 100 - 1 GACCCUGUUCUGGUUUCACUGCAACUGCGCCCAAGAUAUCCAUCCAGUUUAGUUGAAACCAGAGCAGGCUGAC-CCUGCAUUCCCUGCCACGCUUUGAACU ...((((((((((((((((((.(((((.................)))))))).))))))))))))))).....-...(((.....)))............. ( -30.63) >DroYak_CAF1 42767 85 - 1 GAUCCUGUUCUGGUUUCA---------------AGUUCUGGUCUAAGUUUAGUUGAAACCAGAGCAGGCUGAC-CCUGCAUUCCCUGCCACUGCUUGAACU ...(((((((((((((((---------------(.(.((......))...).)))))))))))))))).....-...(((.....)))............. ( -27.90) >consensus GAUCCUGUUCUGGUUUCA_________CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGACCCCUGCAUUCCCUGCCACGCUUUGAAUU ...(((((((((((((((...................................))))))))))))))).........((.......))............. (-20.79 = -20.59 + -0.20)

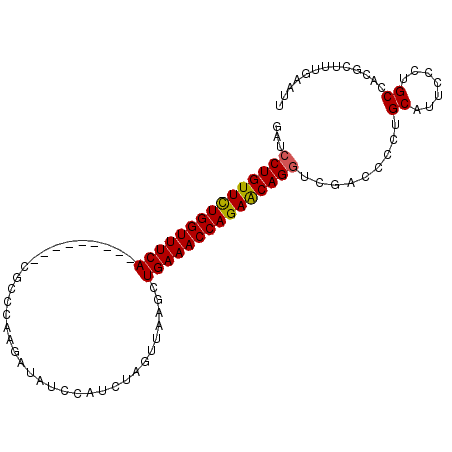
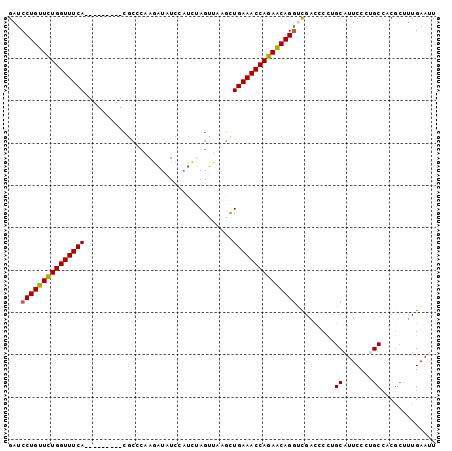
| Location | 1,154,469 – 1,154,560 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -19.59 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1154469 91 + 22407834 AGGGGUCGACCUGUUCUGGUUUCAGCUUAACUAUAUGGAUAUUUUGGGCG---------UGAAACCAAAACAGGAUCUUGGGUAAGCAUACAUGUCGUUC .((.((.((((((((.((((((((((((((..(((....))).)))))).---------)))))))).)))))).))....(((....))))).)).... ( -26.00) >DroSec_CAF1 47985 91 + 1 AGGGGACGACCUGUUCUGGUUUCAGCUUAACUAGAUGGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUCUUGGCUAAGCAUACAUGUCGUUC ...((((((((((((((((((((((((((...((((....))))))))).---------)))))))))))))))......((...)).......)))))) ( -36.70) >DroSim_CAF1 43029 91 + 1 AAGGAUCGAUCUGUUCUGGUUUCAGCUUAACUAGAUUGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUCUUGGCUAAGCAUACAUGUCGUUC .((((((...(((((((((((((((((((...((((....))))))))).---------))))))))))))))))))))(((...........))).... ( -34.60) >DroEre_CAF1 41667 99 + 1 AGG-GUCAGCCUGCUCUGGUUUCAACUAAACUGGAUGGAUAUCUUGGGCGCAGUUGCAGUGAAACCAGAACAGGGUCUGGAGUAAGCAUACAUGUCGUUC ...-.((((((((.((((((((((....((((((..((....))....).)))))....)))))))))).))))..)))).(((....)))......... ( -30.30) >DroYak_CAF1 42791 84 + 1 AGG-GUCAGCCUGCUCUGGUUUCAACUAAACUUAGACCAGAACU---------------UGAAACCAGAACAGGAUCUUGAGUAAGCAUACAUGUCAUUC ..(-.(((.((((.(((((((((((..................)---------------)))))))))).))))....))).)................. ( -21.37) >consensus AGGGGUCGACCUGUUCUGGUUUCAGCUUAACUAGAUGGAUAUCUUGGGCG_________UGAAACCAGAACAGGAUCUUGGGUAAGCAUACAUGUCGUUC .........(((((((((((((((...................................))))))))))))))).......................... (-19.59 = -20.03 + 0.44)

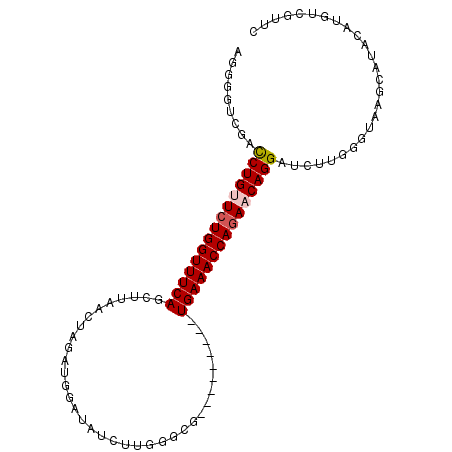
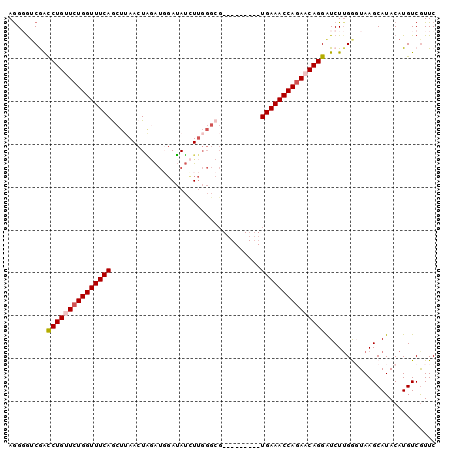
| Location | 1,154,469 – 1,154,560 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -20.63 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1154469 91 - 22407834 GAACGACAUGUAUGCUUACCCAAGAUCCUGUUUUGGUUUCA---------CGCCCAAAAUAUCCAUAUAGUUAAGCUGAAACCAGAACAGGUCGACCCCU ...((((..(((....)))........((((((((((((((---------.((......(((....))).....))))))))))))))))))))...... ( -26.00) >DroSec_CAF1 47985 91 - 1 GAACGACAUGUAUGCUUAGCCAAGAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGUCCCCU ....(((......((...))...((.(((((((((((((((---------.((.(.((((....)))).)....)))))))))))))))))))))).... ( -30.90) >DroSim_CAF1 43029 91 - 1 GAACGACAUGUAUGCUUAGCCAAGAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCAAUCUAGUUAAGCUGAAACCAGAACAGAUCGAUCCUU .......................((((.(((((((((((((---------.((.(.((((....)))).)....)))))))))))))))))))....... ( -27.50) >DroEre_CAF1 41667 99 - 1 GAACGACAUGUAUGCUUACUCCAGACCCUGUUCUGGUUUCACUGCAACUGCGCCCAAGAUAUCCAUCCAGUUUAGUUGAAACCAGAGCAGGCUGAC-CCU .........(((....)))..(((..((((((((((((((((((.(((((.................)))))))).))))))))))))))))))..-... ( -31.93) >DroYak_CAF1 42791 84 - 1 GAAUGACAUGUAUGCUUACUCAAGAUCCUGUUCUGGUUUCA---------------AGUUCUGGUCUAAGUUUAGUUGAAACCAGAGCAGGCUGAC-CCU ...(((...(((....))))))....(((((((((((((((---------------(.(.((......))...).)))))))))))))))).....-... ( -27.30) >consensus GAACGACAUGUAUGCUUACCCAAGAUCCUGUUCUGGUUUCA_________CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGACCCCU ..........................(((((((((((((((...................................)))))))))))))))......... (-20.63 = -20.43 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:03 2006