
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,610,322 – 10,610,458 |
| Length | 136 |
| Max. P | 0.879187 |

| Location | 10,610,322 – 10,610,422 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
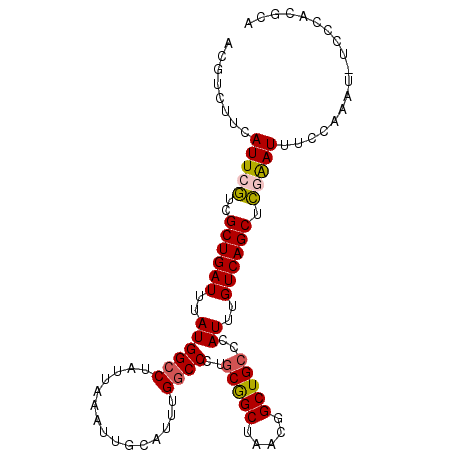
>2L_DroMel_CAF1 10610322 100 + 22407834 ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGACCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU-UCCCACGCA ........(((((..((((((..((((....((((((...))))))....(((((.....)))))))))..)))))).)))))........-......... ( -24.50) >DroSec_CAF1 89920 100 + 1 ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU-UCCCACGCA ........(((((..((((((..((((((...............))))..(((((.....)))))..))..)))))).)))))........-......... ( -25.46) >DroSim_CAF1 90466 100 + 1 ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU-UCCCACGCA ........(((((..((((((..((((((...............))))..(((((.....)))))..))..)))))).)))))........-......... ( -25.46) >DroEre_CAF1 90793 99 + 1 ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCU-CGGCUAACGGCUGCCCAUUUGUCAGCUCUAAUUUCCAAAU-UCCCACGCA (((........))).((((((..((((.(.........((..((((((..-.))))))..)).).))))..))))))..............-......... ( -20.60) >DroYak_CAF1 93228 101 + 1 ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAUUUCCCACGCA ........(((((..((((((..((((((...............))))..(((((.....)))))..))..)))))).))))).................. ( -25.46) >DroAna_CAF1 92399 99 + 1 ACGGCUUUAUUCAUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCAGCGCUCGGCUGC-CAUUUGUCAGCUUAGAUUGCCAGGU-UCCCACGAA ..(((.....((...((((((..((((((...............))))..(((((.....)))))-.))..))))))...))..)))....-......... ( -27.06) >consensus ACGUCUUCAUUCGUCGCUGAUUUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU_UCCCACGCA ........(((((..((((((..((((((...............))))..(((((.....)))))..))..)))))).))))).................. (-21.71 = -21.83 + 0.11)


| Location | 10,610,343 – 10,610,458 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -18.68 |
| Energy contribution | -17.47 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.581018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
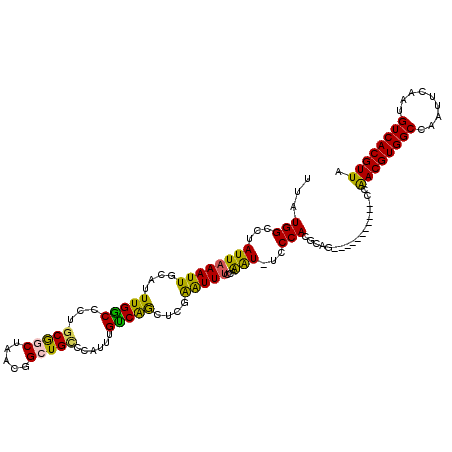
>2L_DroMel_CAF1 10610343 115 + 22407834 UUAUGGCCUAUUAAAUUGCAUUUGACCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU-UCCCACGCAGCCAGCU----AACAACGUGGCCAAUUCAAUGUCACGUUA ...((((.........(((..(((((...(((((.....)))))......)))))...(((((....)))-))....)))))))...----...((((((((.........)))))))). ( -26.90) >DroPse_CAF1 91063 101 + 1 UUAUGGCCUAUUAAAUUGCAGUUGGCCCUGCGG--------UGUUCUGUGGCCGACUCGAGUUUCGAAAUUUCCCACGGCA-----------CAGACGUGGCCAAUUCAGUGUCACGUUA ...(((((............(((((((..((((--------....)))))))))))(((.....)))..........))).-----------))(((((((((......).)))))))). ( -32.70) >DroEre_CAF1 90814 111 + 1 UUAUGGCCUAUUAAAUUGCAUUUGGCCCU-CGGCUAACGGCUGCCCAUUUGUCAGCUCUAAUUUCCAAAU-UCCCACGCAGCC-------GCUCAACGUGGCCAAUUCAAUGUCACGUUA ...((((.......((((...((((((..-((.....(((((((..(((((..((......))..)))))-......))))))-------).....)).))))))..))))))))..... ( -27.01) >DroYak_CAF1 93249 120 + 1 UUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAUUUCCCACGCAGCCAGCCUUUGGCCAACGUGGCCAAUUCAAUGUCACGUUA ....((((.........(((........)))((((...((((((..(((((......)))))...............))))))))))...))))((((((((.........)))))))). ( -33.83) >DroAna_CAF1 92420 107 + 1 UUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCAGCGCUCGGCUGC-CAUUUGUCAGCUUAGAUUGCCAGGU-UCCCACGAAC-----------GGAACGUGGCCAAUUCAGUGUCACGUUA ....((((...............))))..(((((.....)))))-.....((..(((..((((((((.((-(((.......-----------))))).))).))))).)))...)).... ( -30.76) >DroPer_CAF1 92981 101 + 1 UUAUGGCCUAUUAAAUUGCAGUUGGCCCUGCGG--------UGUUCUGUGGCCGACUCGAGUUUCGAAAUUUCCCACGGCA-----------CAGACGUGGCCAAUUCAGUGUCACGUUA ...(((((............(((((((..((((--------....)))))))))))(((.....)))..........))).-----------))(((((((((......).)))))))). ( -32.70) >consensus UUAUGGCCUAUUAAAUUGCAUUUGGCCCUGCGGCUAACGGCUGCCCAUUUGUCAGCUCGAAUUUCCAAAU_UCCCACGCAG___________CCAACGUGGCCAAUUCAAUGUCACGUUA ...(((...((((((((....(((((...(((((.....)))))......)))))....)))))...)))...)))..................((((((((.........)))))))). (-18.68 = -17.47 + -1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:39 2006