
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,605,356 – 10,605,474 |
| Length | 118 |
| Max. P | 0.741437 |

| Location | 10,605,356 – 10,605,474 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
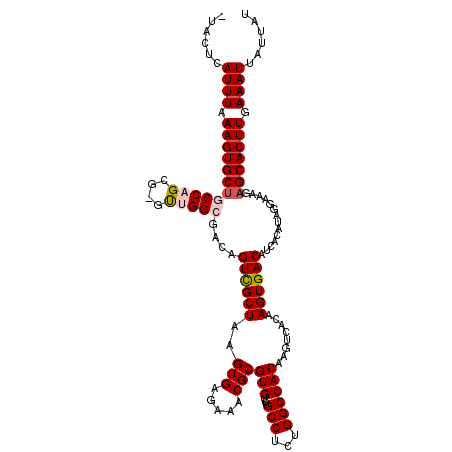
>2L_DroMel_CAF1 10605356 118 + 22407834 AUAAUAAUUUCAAGUGCUCUUUCCUAUGUGAUGUCACUUGUGACUUGUGGCCAGAGGCAGACACGCGUUUCUCACUUAGCGACUGUCGCCAAC-CGCUGGCAGCACUUUAAAUGACUAA ......((((.(((((((..............((((....)))).....(((((.(((.((((..((((........))))..)))))))...-..)))))))))))).))))...... ( -34.60) >DroSec_CAF1 84989 118 + 1 AUAAUAAUUUCAAGUGCUCUUUCCUAUGUGAUGUCACUUGUGACUUGUGGCCAGAGGCAGACACGCGUUUCUCACUUAGCGACUGUCGCCAGCGCGCUGGCAGCACUUUAAAUGAGUA- ......((((.(((((((....(((..((.((((((....))))..)).))...)))..((((..((((........))))..))))(((((....)))))))))))).)))).....- ( -34.60) >DroSim_CAF1 85545 118 + 1 AUAUUAAUUUCAAGUGCUCUUUCCUAUGUGAUGUCACUUGUGACUUGUGGCCACAGGCAGACACGCGUUUCUCACUUAGCGACUGUCGCCAGCGCGCUGGCAGCACUUUAAAUGAGUA- .((((.((((.(((((((........((((..(((((.........)))))))))(((((...(((............))).)))))(((((....)))))))))))).)))).))))- ( -36.80) >DroEre_CAF1 86008 105 + 1 AUAAUAAUUUCAAGUGCUC-----UAUGUAGUGUCACU-------UGUGGCCAAAGGCAGACACGCGUUUCCCACUUAGCAACUGUCGCCGAG-CGCUGGGCGCACUUUAAAUGAGUU- ......((((.((((((..-----.((((.(((((.(.-------...)(((...))).)))))))))..((((((..((.......))..))-...)))).)))))).)))).....- ( -26.60) >DroYak_CAF1 88155 110 + 1 AUAAUAAUUUCAAGUGCUCUCUCCUAUGUGAUGUCACU-------UGUGGCCAAAGGCAGACACGCGUUUCUCACUUAGCGACUGUCGCCGAC-UGCUGGGAGCACUUUAAAUGAGUU- ......((((.((((((((((......(((....))).-------...(((....(((.((((..((((........))))..)))))))...-.))))))))))))).)))).....- ( -35.10) >consensus AUAAUAAUUUCAAGUGCUCUUUCCUAUGUGAUGUCACUUGUGACUUGUGGCCAAAGGCAGACACGCGUUUCUCACUUAGCGACUGUCGCCAAC_CGCUGGCAGCACUUUAAAUGAGUA_ ......((((.(((((((....((...(((..(((((.........)))))....(((.((((..((((........))))..)))))))....))).)).))))))).))))...... (-26.54 = -27.42 + 0.88)



| Location | 10,605,356 – 10,605,474 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10605356 118 - 22407834 UUAGUCAUUUAAAGUGCUGCCAGCG-GUUGGCGACAGUCGCUAAGUGAGAAACGCGUGUCUGCCUCUGGCCACAAGUCACAAGUGACAUCACAUAGGAAAGAGCACUUGAAAUUAUUAU .((((.((((.(((((((.((((((-..((....))..))))..((((.......(((...(((...))))))..((((....)))).))))...))....))))))).)))).)))). ( -37.70) >DroSec_CAF1 84989 118 - 1 -UACUCAUUUAAAGUGCUGCCAGCGCGCUGGCGACAGUCGCUAAGUGAGAAACGCGUGUCUGCCUCUGGCCACAAGUCACAAGUGACAUCACAUAGGAAAGAGCACUUGAAAUUAUUAU -.....((((.(((((((.((((((.((((....))))))))..((((.......(((...(((...))))))..((((....)))).))))...))....))))))).))))...... ( -35.90) >DroSim_CAF1 85545 118 - 1 -UACUCAUUUAAAGUGCUGCCAGCGCGCUGGCGACAGUCGCUAAGUGAGAAACGCGUGUCUGCCUGUGGCCACAAGUCACAAGUGACAUCACAUAGGAAAGAGCACUUGAAAUUAAUAU -.....((((.(((((((..(((((((((((((.....))))).((.....))))))).)))((((((.......((((....))))....))))))....))))))).))))...... ( -37.50) >DroEre_CAF1 86008 105 - 1 -AACUCAUUUAAAGUGCGCCCAGCG-CUCGGCGACAGUUGCUAAGUGGGAAACGCGUGUCUGCCUUUGGCCACA-------AGUGACACUACAUA-----GAGCACUUGAAAUUAUUAU -.....((((.((((((((....((-((..((((...))))..))))(....)))(((((....((((....))-------)).)))))......-----..)))))).))))...... ( -28.50) >DroYak_CAF1 88155 110 - 1 -AACUCAUUUAAAGUGCUCCCAGCA-GUCGGCGACAGUCGCUAAGUGAGAAACGCGUGUCUGCCUUUGGCCACA-------AGUGACAUCACAUAGGAGAGAGCACUUGAAAUUAUUAU -.....((((.((((((((((...(-(.((((....))))))..((((....(((.(((..(((...))).)))-------.)))...))))...))...)))))))).))))...... ( -30.30) >consensus _UACUCAUUUAAAGUGCUGCCAGCG_GUUGGCGACAGUCGCUAAGUGAGAAACGCGUGUCUGCCUCUGGCCACAAGUCACAAGUGACAUCACAUAGGAAAGAGCACUUGAAAUUAUUAU ......((((.((((((((((((....)))))....((((((..(((.....)))(((...(((...))))))........))))))..............))))))).))))...... (-26.30 = -26.90 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:34 2006