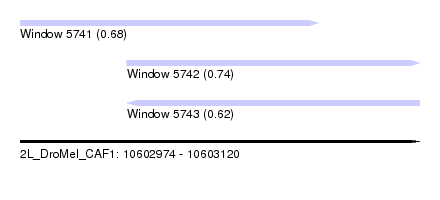
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,602,974 – 10,603,120 |
| Length | 146 |
| Max. P | 0.741842 |

| Location | 10,602,974 – 10,603,083 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
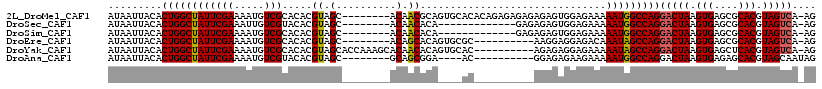
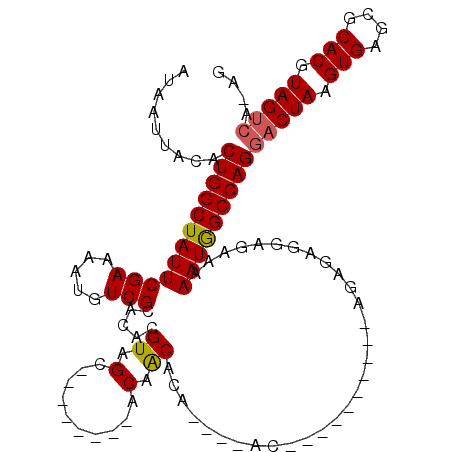
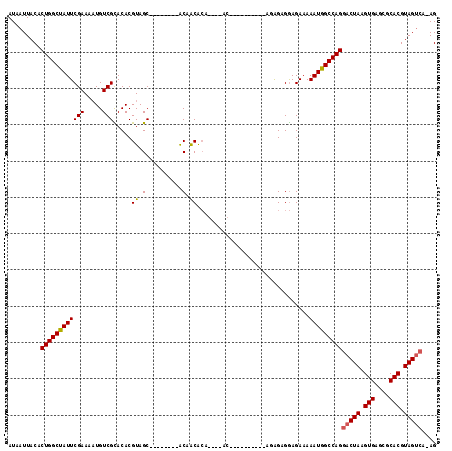
>2L_DroMel_CAF1 10602974 109 + 22407834 AUAAUUACACUGGCUAUUCGAAAAUGUCGCACACGUAGC--------ACAACGCAGUGCACACAGAGAGAGAGAGUGGAGAAAAAUGGCCAGGACUAAGUGAGCGCACGUAGUCA-AG .........(((((((((..........((((.(((...--------...)))..)))).(((...........)))......)))))))))(((((.(((....))).))))).-.. ( -30.00) >DroSec_CAF1 82685 96 + 1 AUAAUUACACUGGCUAUUCGAAAUUGUCGUACACGUAGC--------ACAACACA-------------GAGAGAGUGGAGAAAAAUGGCCAGGACUAAGUGAGCGCACGUAGUCA-AG .........(((((((((.....((((.((.......))--------))))(((.-------------......)))......)))))))))(((((.(((....))).))))).-.. ( -24.30) >DroSim_CAF1 83244 96 + 1 AUAAUUACACUGGCUAUUCGAAAAUGUCGCACACGUAGC--------ACAACACA-------------GAGAGAGUGGAGAAAAAUGGCCAGGACUAAGUGAGCGCACGUAGUCA-AG .........(((((((((......(((..(....)..))--------)(..(((.-------------......)))..)...)))))))))(((((.(((....))).))))).-.. ( -25.80) >DroEre_CAF1 83553 99 + 1 AUAAUUACACUGGCUAUUCGAAAAUGUCGCACACGUAGC--------ACAGCACAGUGCGC----------AAGGAGGAGACAAAUAGCCAGGACUAAGUGAGCGCACGUAGUCA-AG .........(((((((((......((.(((((..((.((--------...)))).))))))----------)....(....).)))))))))(((((.(((....))).))))).-.. ( -32.10) >DroYak_CAF1 85761 107 + 1 AUAAUUACACUGGCUAUUCGAAAAUGUCGCACACGUAGCACCAAAGCACAACACAGUGCAC----------AGAGAGGAGAAAAAUAGCCAGGACUAAGUGAGCUCACGUAGUCA-AG .........(((((((((......(((..(....)..))).....((((......))))..----------............)))))))))(((((.(((....))).))))).-.. ( -30.80) >DroAna_CAF1 85228 96 + 1 AUAAUUACACUGGCUAUUCGAAAAUGUCGUACACGUAGC--------GCAGCGGA----AC----------GGAGAGAAGAAAAAUGGCCAGGACUAAGUGAGAGCACGUAGCAAUAG ....((((.(((((((((........((((.(.(((...--------...)))).----))----------))..........)))))))))......((....))..))))...... ( -21.77) >consensus AUAAUUACACUGGCUAUUCGAAAAUGUCGCACACGUAGC________ACAACACA____AC__________AGAGAGGAGAAAAAUGGCCAGGACUAAGUGAGCGCACGUAGUCA_AG .........((((((((((((.....))).....((.(..........).))...............................)))))))))(((((.(((....))).))))).... (-18.99 = -18.88 + -0.11)
| Location | 10,603,013 – 10,603,120 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10603013 107 + 22407834 -------ACAACGCAGUGCACACAGAGAGAGAGAGUGGAGAAAAAUGG-CCAGGACUAAGUGA-GCGCACGUAGUCAAGGAAAUAAUACACAUACGCACUGUGGGCCGUUGAGA-AA -------....((((((((.(((...........))).......((..-((..(((((.(((.-...))).)))))..))..))...........))))))))...........-.. ( -27.80) >DroSec_CAF1 82724 94 + 1 -------ACAACACA-------------GAGAGAGUGGAGAAAAAUGG-CCAGGACUAAGUGA-GCGCACGUAGUCAAGGAAAUAAUACACAUACGCACUGUGGGCCAUUGAGA-AA -------.(..(((.-------------......)))..)...(((((-((..(((((.(((.-...))).)))))............((((.......)))))))))))....-.. ( -24.70) >DroSim_CAF1 83283 94 + 1 -------ACAACACA-------------GAGAGAGUGGAGAAAAAUGG-CCAGGACUAAGUGA-GCGCACGUAGUCAAGGAAAUAAUACACAUACGCACUGUGGGCCAUUGAGA-AA -------.(..(((.-------------......)))..)...(((((-((..(((((.(((.-...))).)))))............((((.......)))))))))))....-.. ( -24.70) >DroEre_CAF1 83592 97 + 1 -------ACAGCACAGUGCGC----------AAGGAGGAGACAAAUAG-CCAGGACUAAGUGA-GCGCACGUAGUCAAGGAAAUAAUACACAUACGCACUGUGGGCCGUUGAGA-AA -------....(((((((((.----------.....(....)......-((..(((((.(((.-...))).)))))..))..............)))))))))...........-.. ( -29.30) >DroYak_CAF1 85801 104 + 1 CCAAAGCACAACACAGUGCAC----------AGAGAGGAGAAAAAUAG-CCAGGACUAAGUGA-GCUCACGUAGUCAAGCAAAUAAUACACAUACGCACUGUGGGCCGUUGCGA-AA .....((((..((((((((..----------.....((..(....)..-))..(((((.(((.-...))).)))))...................))))))))....).)))..-.. ( -27.10) >DroPer_CAF1 86541 95 + 1 --------CGGGGCU-------------GA-GGAGAGAAGAAAAAUUGCCCAGGACUAAGUGGGGAGCACGUAGCAGAGGAAAUAAUACACAUACGCACUGUGAGCAGUGGAGAAAA --------..((((.-------------..-................))))..(.(((.(((.....))).)))).....................(((((....)))))....... ( -16.81) >consensus _______ACAACACA_____________GA_AGAGAGGAGAAAAAUGG_CCAGGACUAAGUGA_GCGCACGUAGUCAAGGAAAUAAUACACAUACGCACUGUGGGCCGUUGAGA_AA ...........................................(((((.((..(((((.(((.....))).)))))............((((.......)))))))))))....... (-14.07 = -14.68 + 0.61)
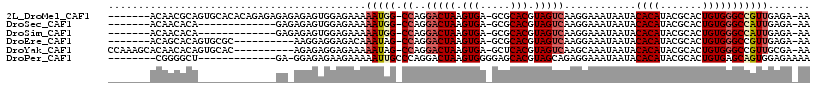
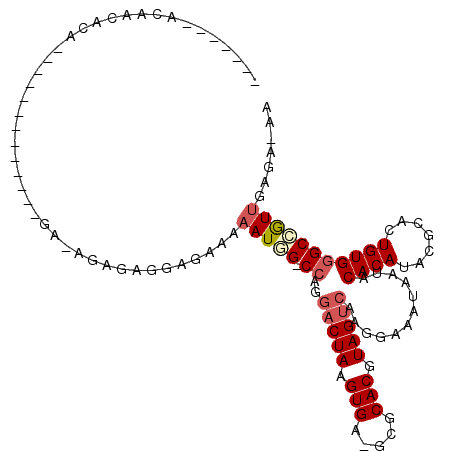
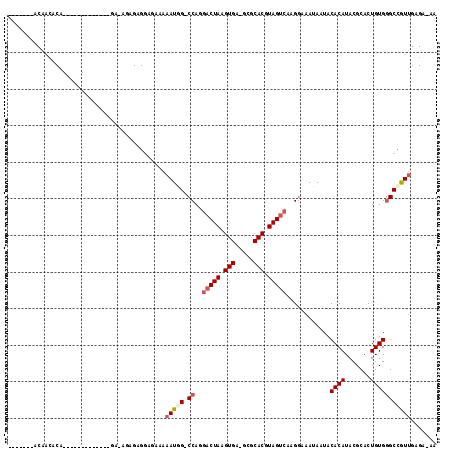
| Location | 10,603,013 – 10,603,120 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10603013 107 - 22407834 UU-UCUCAACGGCCCACAGUGCGUAUGUGUAUUAUUUCCUUGACUACGUGCGC-UCACUUAGUCCUGG-CCAUUUUUCUCCACUCUCUCUCUCUGUGUGCACUGCGUUGU------- ..-...(((((.....(((((((..............((..(((((.(((...-.))).)))))..))-...........(((...........))))))))))))))).------- ( -23.30) >DroSec_CAF1 82724 94 - 1 UU-UCUCAAUGGCCCACAGUGCGUAUGUGUAUUAUUUCCUUGACUACGUGCGC-UCACUUAGUCCUGG-CCAUUUUUCUCCACUCUCUC-------------UGUGUUGU------- ..-....(((((((.((((((((((((((..(((......))).)))))))))-..)))..))...))-)))))......(((......-------------.)))....------- ( -21.10) >DroSim_CAF1 83283 94 - 1 UU-UCUCAAUGGCCCACAGUGCGUAUGUGUAUUAUUUCCUUGACUACGUGCGC-UCACUUAGUCCUGG-CCAUUUUUCUCCACUCUCUC-------------UGUGUUGU------- ..-....(((((((.((((((((((((((..(((......))).)))))))))-..)))..))...))-)))))......(((......-------------.)))....------- ( -21.10) >DroEre_CAF1 83592 97 - 1 UU-UCUCAACGGCCCACAGUGCGUAUGUGUAUUAUUUCCUUGACUACGUGCGC-UCACUUAGUCCUGG-CUAUUUGUCUCCUCCUU----------GCGCACUGUGCUGU------- ..-.....(((((..((((((((((................(((((.(((...-.))).)))))..((-(.....))).......)----------))))))))))))))------- ( -29.80) >DroYak_CAF1 85801 104 - 1 UU-UCGCAACGGCCCACAGUGCGUAUGUGUAUUAUUUGCUUGACUACGUGAGC-UCACUUAGUCCUGG-CUAUUUUUCUCCUCUCU----------GUGCACUGUGUUGUGCUUUGG ..-..(((.((((..((((((((((.......)))..(((.(((((.(((...-.))).)))))..))-)................----------..))))))))))))))..... ( -27.80) >DroPer_CAF1 86541 95 - 1 UUUUCUCCACUGCUCACAGUGCGUAUGUGUAUUAUUUCCUCUGCUACGUGCUCCCCACUUAGUCCUGGGCAAUUUUUCUUCUCUCC-UC-------------AGCCCCG-------- .......(((((....))))).(((((((((..........))).))))))...............((((................-..-------------.))))..-------- ( -15.41) >consensus UU_UCUCAACGGCCCACAGUGCGUAUGUGUAUUAUUUCCUUGACUACGUGCGC_UCACUUAGUCCUGG_CCAUUUUUCUCCACUCU_UC_____________UGUGUUGU_______ ..........((.(((.((((((....))))))........(((((.(((.....))).))))).))).)).............................................. (-12.72 = -12.17 + -0.55)

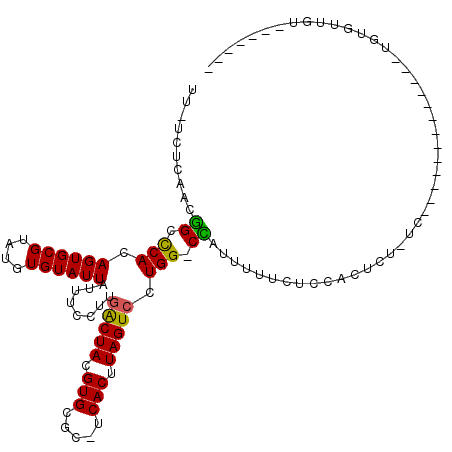
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:30 2006