| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
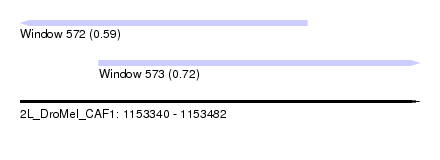
| Location | 1,153,340 – 1,153,482 |
| Length | 142 |
| Max. P | 0.721787 |

| Location | 1,153,340 – 1,153,442 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.35 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
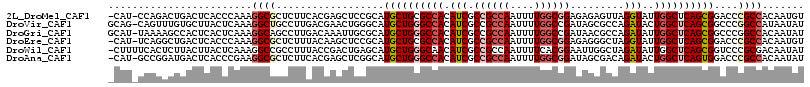
>2L_DroMel_CAF1 1153340 102 - 22407834 CGCCAAAAUUGGCGGCGAUGUGGCGCAGCAUGCGGAGCUCGUGAAGAGCGCCUUUGGGUGAGUCAGUCUGGAUGAGUCAUCUAUGGUAAAAGUGUAAUAAUG (((((....)))))(((......))).((((.....((((.....))))(((..(((((((.(((.......))).))))))).)))....))))....... ( -34.30) >DroSec_CAF1 46879 101 - 1 CGCCAAAAUUGGCGGAGAUGUGGCGCAGCAUGCGGAGCUCGUCAGGAGCGCCUUUGGGUGAGUCAGUCUGGAUGAGUCAUCUAUGGGUAAAGUGUAAU-AUG (((((....)))))...((((.((((..........((((.....))))((((.(((((((.(((.......))).))))))).))))...)))).))-)). ( -36.40) >DroSim_CAF1 41926 102 - 1 CGUCAAAAUUGGCGGCGAUGUGGCGCAGCAUGCGGAGCUCGUGAGGAGCGCCUUUGGGUGAGUCAUUUUGGAUGAGUCAUCUAUGGGUAAAGUGUAAUAAUG (((((....)))))(((......))).((((.....((((.....))))((((.(((((((.(((((...))))).))))))).))))...))))....... ( -34.40) >DroEre_CAF1 40586 102 - 1 CGCCAAAAUUGGCGGCGAUGUGGCGCAGCAUGCGGAGCUUGUAAAGAGCGCCUUUGGGUGAGUCAGCCUGAAUGAGUCAUCUAUGGGAGAAGUGGAAUAAUG (((((....)))))((((.((..(((.....)))..)))))).....((.(((.(((((((.(((.......))).))))))).)))....))......... ( -31.30) >DroYak_CAF1 41676 102 - 1 CGCCAAAAUUGGCGGCGAUGUGGCGCAGCAUGCGGAGCUCGUAAAGAGCGCCUUUGGGUGAGUCAGUCUGGAUGAGUCAUCCGUGCGAAAAGUGAAAUAAUG (((((....)))))(((......))).((((..((.((((.....)))).))...((((((.(((.......))).))))))))))................ ( -37.20) >DroAna_CAF1 46301 98 - 1 CGCCAAAAUUGGCGGCGAUGUGGCCCAGCAUGCCGAGCUCGUGAAGAGCGCCUUCGGGUGAGUCAUCCGGCAUGAGUAAUCCAU---UAAAGUAUCC-ACUU (((((....)))))((...((((.....(((((((.((((.....))))(((....)))........)))))))......))))---....))....-.... ( -36.60) >consensus CGCCAAAAUUGGCGGCGAUGUGGCGCAGCAUGCGGAGCUCGUGAAGAGCGCCUUUGGGUGAGUCAGUCUGGAUGAGUCAUCUAUGGGAAAAGUGUAAUAAUG (((((....))))).......(((((.....))...((((.....)))))))..(((((((.(((.......))).)))))))................... (-28.46 = -28.35 + -0.11)

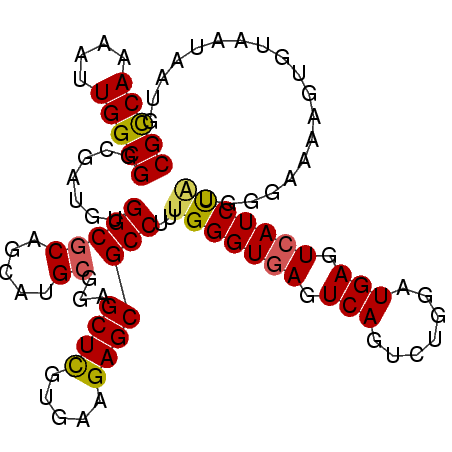
| Location | 1,153,368 – 1,153,482 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -22.69 |
| Energy contribution | -23.83 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1153368 114 + 22407834 -CAU-CCAGACUGACUCACCCAAAGGCGCUCUUCACGAGCUCCGCAUGCUGCGCCACAUCGCCGCCAAUUUUGGCGGAGAGAGUUAGGUAUUGGCUCAGCGGACCCGCCACAAUGU -...-((((.(((((((.......(((((((.....))))...((.....)))))...((.((((((....)))))).)))))))))...))))....(((....)))........ ( -41.80) >DroVir_CAF1 53071 115 + 1 GCAG-CAGUUUGUGCUUACUCAAAGGCUGCCUUGACGAACUGGGCAUGCUGGGCCACAUCGCCGCCAAUUUUGGCCGAUAGCGCCAGAUACUGGCUCAGCGGCCCGGCCAUAAUAU ((((-(..((((........)))).))))).........((((((.((((((((......)))((((..(((((((....).))))))...)))).)))))))))))......... ( -46.50) >DroGri_CAF1 45014 115 + 1 GCAU-UAAAAGCCACUCACUCAAAGGCAGCCUUGACAAAUUGCGCAUGCUGGGCCACAUCGCCGCCAAUUUUGGCCGAUAACGCCAGAUAUUGGCUCAGCGGCCCGGCCACAAUAU ((..-.....(((...........))).((..(.....)..))))..((((((((.....((.((((((((((((.......)))))).))))))...))))))))))........ ( -37.40) >DroEre_CAF1 40614 114 + 1 -CAU-UCAGGCUGACUCACCCAAAGGCGCUCUUUACAAGCUCCGCAUGCUGCGCCACAUCGCCGCCAAUUUUGGCGGAGAGGGCUAGGUAUUGGCUCAGCGGACCCGCCACAAUGU -...-....(((((..((......((.(((.......))).))..(((((..(((...((.((((((....)))))).)).)))..)))))))..)))))((.....))....... ( -37.10) >DroWil_CAF1 79095 115 + 1 -CUUUUCACUCUUACUUACUCAAAGGCCGCCUUUACCGACUGAGCAUGCUGGGCAACAUCGCCGCCAAUUUUCACGGAAUUGGCUAGAUAUUGGCUCAGCGGUCCCGCGACAAUAU -..........................(((....((((.((((((...((((((......)))((((((((.....)))))))))))......))))))))))...)))....... ( -34.60) >DroAna_CAF1 46325 114 + 1 -CAU-GCCGGAUGACUCACCCGAAGGCGCUCUUCACGAGCUCGGCAUGCUGGGCCACAUCGCCGCCAAUUUUGGCGGAUAGCGACAGAUACUGGCUCAGUGGACCCGCCACAAUAU -(((-(((((........((....)).((((.....)))))))))))).(((((((.(((.((((((....)))))).........)))..)))))))((((.....))))..... ( -42.70) >consensus _CAU_UCAGACUGACUCACCCAAAGGCGCCCUUCACGAACUCCGCAUGCUGGGCCACAUCGCCGCCAAUUUUGGCGGAUAGCGCCAGAUAUUGGCUCAGCGGACCCGCCACAAUAU ........................((((..................((((((((((.(((.((((((....)))))).........)))..))))))))))....))))....... (-22.69 = -23.83 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:59 2006