
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,591,985 – 10,592,148 |
| Length | 163 |
| Max. P | 0.865915 |

| Location | 10,591,985 – 10,592,088 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
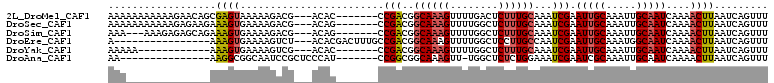
>2L_DroMel_CAF1 10591985 103 - 22407834 CAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA----------CUG----CCUUACCCCACUGCUUCCAUCUACAUCCCUAUGUA---GUACAUA ....((((.((((.....)))))).(((((.....))))).))(((((....))))).----------...----....................((((((....)))))---)...... ( -15.90) >DroPse_CAF1 81784 104 - 1 AAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUACAGGCUUAGAAGAGCCC----CAUCUUCUGCCCCCU-CCCCCGGCCACUAUCCUCUCCAU------U-----UCCCCUA ......((.((((.....)))))).(((((.....)))))...(((((((((((....----..)))))))......-.....))))..............------.-----....... ( -16.70) >DroSec_CAF1 79295 104 - 1 CAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA----------CUGU---CCUUACCCCACAGCUUCCAUCUACAUCCUUAUGUA---GUACAUA ......((.((((.....))))))...........((..(((.(((((....))))).----------))).---.)).................((((((....)))))---)...... ( -17.50) >DroSim_CAF1 79876 114 - 1 CAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCACUGCCACCCACUGU---CCUUACCCCACAGCUUCCAUCUACAUCCUUAUGUA---GUACAUA ....((((.((((.....)))))).(((((.....))))).))(((((....)))))...........((((---.........)))).......((((((....)))))---)...... ( -16.90) >DroEre_CAF1 80291 107 - 1 CCAAUCGAAUUGCAAAUGGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGUCAAGCCA----------CUAUCCGCUUCACCCCACAACUUCCAUCUGCCUCCCUGUGUA---GUACAUA ......(.((((((...((((....(((((.....))))).(((((.((((......)----------)))...))))).................))))......))))---)).)... ( -19.40) >DroYak_CAF1 82303 110 - 1 CAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCG----------CUGCCCUCUUCACCCCACAACUUCCAUCUACAUCCCUAUGUAGUAGUACAUA ......((.((((.....))))))...........((..(((.(((((....))))).----------)))..))............(((.....((((((....)))))).)))..... ( -18.00) >consensus CAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA__________CUGU___CCUCACCCCACAACUUCCAUCUACAUCCCUAUGUA___GUACAUA ......((.((((.....)))))).(((((.....)))))...(((((....)))))............................................................... (-11.72 = -11.58 + -0.14)

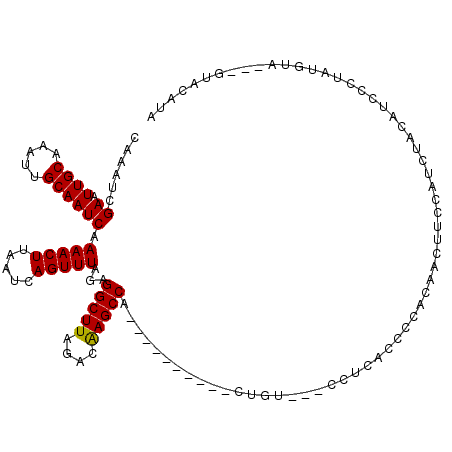
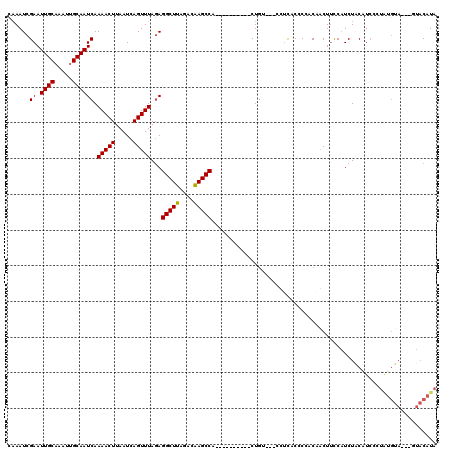
| Location | 10,592,022 – 10,592,118 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.765883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10592022 96 + 22407834 UAAGG----CAG----------UGGCUUGUCUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGUCAAAACUUUGCCGUCGG-------GUGU---C ...((----(((----------.(((((....))))).))...(((((.(((((((((((....(((((((......))))))).)))))))))))....)))))-------..))---) ( -30.40) >DroSec_CAF1 79332 97 + 1 UAAGG---ACAG----------UGGCUUGUCUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------CUGU---C ....(---((((----------.(((((....)))))......(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))-------))))---) ( -30.10) >DroSim_CAF1 79913 107 + 1 UAAGG---ACAGUGGGUGGCAGUGGCUUGUCUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------CUGU---C ....(---(((((.(..(((((.(((((....))))).)).........((((((((...((..(((((((......)))))))..)))))))))).)))..).)-------))))---) ( -40.90) >DroEre_CAF1 80328 107 + 1 UGAAGCGGAUAG----------UGGCUUGACUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCCAUUUGCAAUUCGAUUGGCAAGGAGCCAAAACUUUGCCGUCGGCAAAGUCGUGU---A ....(((.....----------((((((.........((.(((((.....))))).))((((((.(((.....))).)))))).))))))..((((((((...)))))))))))..---. ( -32.20) >DroYak_CAF1 82343 100 + 1 UGAAGAGGGCAG----------CGGCUUGUCUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------GUGU---C ....((.(((((----------.(((((....))))).)).........((((((((...((..(((((((......)))))))..)))))))))).))).))..-------....---. ( -31.20) >DroAna_CAF1 81883 96 + 1 ---AGAGGG-------------GAGACCGACUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCGAUUCGAUUUCCAGAGAGCCA-AACUUUGCCGCCGG-------AUGGGAGC ---((((((-------------....))......(((((((((((.....))))).((((((.....))))))........)))).))..-..))))((..((..-------..))..)) ( -20.10) >consensus UAAAG___ACAG__________UGGCUUGUCUAAGCCUCUAAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG_______GUGU___C .......................(((((....)))))......(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))............... (-19.54 = -20.35 + 0.81)


| Location | 10,592,022 – 10,592,118 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10592022 96 - 22407834 G---ACAC-------CCGACGGCAAAGUUUUGACUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA----------CUG----CCUUA .---....-------.....((((((((((((((..(((((((......)))))))..)...)))))))))............(((((....))))).----------.))----))... ( -25.90) >DroSec_CAF1 79332 97 - 1 G---ACAG-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA----------CUGU---CCUUA (---((((-------.....((..((((((((((..(((((((......)))))))..))...))))))))..))........(((((....))))).----------))))---).... ( -27.70) >DroSim_CAF1 79913 107 - 1 G---ACAG-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCACUGCCACCCACUGU---CCUUA (---((((-------..(..((((((((((((((..(((((((......)))))))..))...))))))))............(((((....)))))..))))..)..))))---).... ( -33.10) >DroEre_CAF1 80328 107 - 1 U---ACACGACUUUGCCGACGGCAAAGUUUUGGCUCCUUGCCAAUCGAAUUGCAAAUGGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGUCAAGCCA----------CUAUCCGCUUCA .---....(((((((((...)))))))))(((((.....)))))..((.((((.....))))))............(..(((.(((((....))))).----------)))..)...... ( -30.20) >DroYak_CAF1 82343 100 - 1 G---ACAC-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCG----------CUGCCCUCUUCA .---....-------..((.((((((((((((((..(((((((......)))))))..))...))))))))............(((((....))))).----------.)))).)).... ( -28.70) >DroAna_CAF1 81883 96 - 1 GCUCCCAU-------CCGGCGGCAAAGUU-UGGCUCUCUGGAAAUCGAAUCGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGUCGGUCUC-------------CCCUCU--- (((.((..-------..)).)))......-.........(((.(((((...((.....))..((.(((((.....))))).)).......))))).))-------------).....--- ( -17.80) >consensus G___ACAC_______CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUUAGAGGCUUAGACAAGCCA__________CUGU___CCUUA ................(((..((((((........))))))...)))..((((.....))))((.(((((.....))))).))(((((....)))))....................... (-17.00 = -17.28 + 0.28)


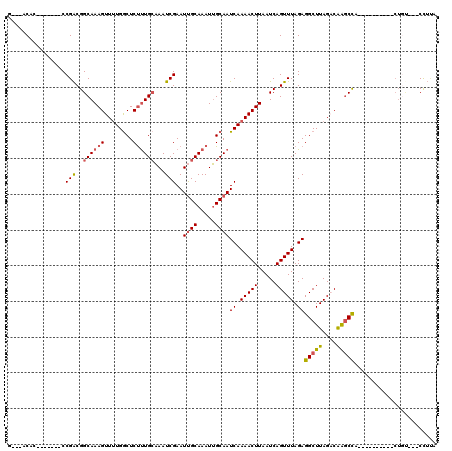
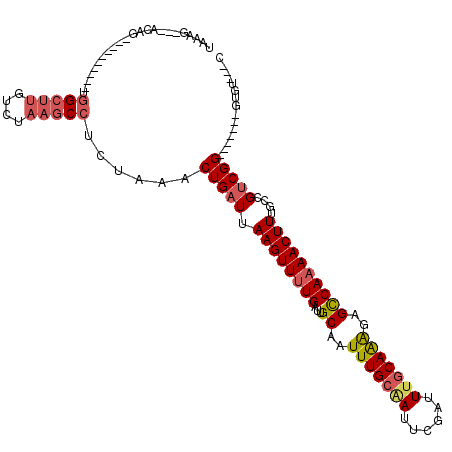
| Location | 10,592,048 – 10,592,148 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10592048 100 + 22407834 AAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGUCAAAACUUUGCCGUCGG-------GUGU---CGUCUUUUUACUCGCUGUUCUUUUUUUUUUU ...(((((.(((((((((((....(((((((......))))))).)))))))))))....)))))-------(((.---.((......)).)))................ ( -22.10) >DroSec_CAF1 79359 100 + 1 AAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------CUGU---CGUCUUUUCACUUUCUUCUCUUUUUUUUUUU ...(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))-------....---............................... ( -19.60) >DroSim_CAF1 79950 97 + 1 AAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------CUGU---CGUCUUUUCACUUUCUGCUCUCUUU---UUU ...(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))-------....---.........................---... ( -19.60) >DroEre_CAF1 80358 91 + 1 AAACUGAUUAAGUUUUGAUUGCCAUUUGCAAUUCGAUUGGCAAGGAGCCAAAACUUUGCCGUCGGCAAAGUCGUGU---AGACUUUUCACUUU----------------U (((((.....))))).((((((.....))))))((((.(((((((........)))))))))))(.((((((....---.)))))).).....----------------. ( -24.10) >DroYak_CAF1 82373 88 + 1 AAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG-------GUGU---CGACUUUUCACUUU------------UUUUU ...(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))-------(((.---........)))...------------..... ( -21.10) >DroAna_CAF1 81907 87 + 1 AAACUGAUUAAGUUUUGAUUGCAAUUUGCGAUUCGAUUUCCAGAGAGCCA-AACUUUGCCGCCGG-------AUGGGAGCGGAUUGCCGCCUU---------------UU (((((.....)))))............((((((((.((((((..(.((..-.........)))..-------.))))))))))))))......---------------.. ( -18.60) >consensus AAACUGAUUAAGUUUUGAUUGCAAUUUGCAAUUCGAUUUGCAAAGAGCCAAAACUUUGCCGUCGG_______GUGU___CGUCUUUUCACUUU______________UUU ...(((((.((((((((...((..(((((((......)))))))..))))))))))....)))))............................................. (-16.59 = -16.73 + 0.14)

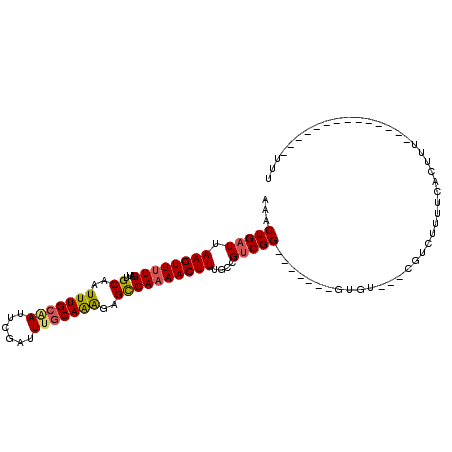
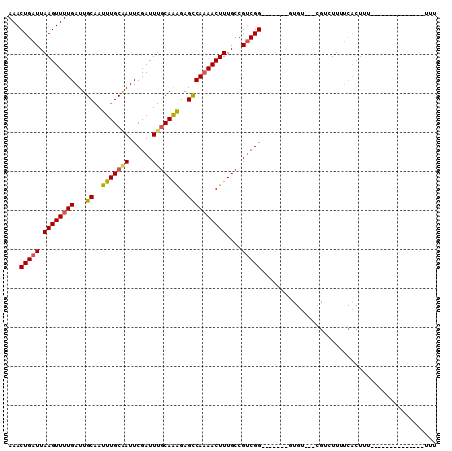
| Location | 10,592,048 – 10,592,148 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
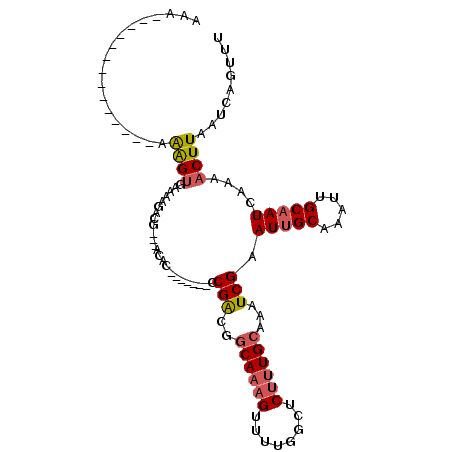
>2L_DroMel_CAF1 10592048 100 - 22407834 AAAAAAAAAAAGAACAGCGAGUAAAAAGACG---ACAC-------CCGACGGCAAAGUUUUGACUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUU ................((((((...(((((.---...(-------(....))....))))).))))(((((((......)))))))..)).....(((((.....))))) ( -16.90) >DroSec_CAF1 79359 100 - 1 AAAAAAAAAAAGAGAAGAAAGUGAAAAGACG---ACAG-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUU .................(((.(((.......---...(-------((...))).((((((((((..(((((((......)))))))..))...))))))))..))).))) ( -20.50) >DroSim_CAF1 79950 97 - 1 AAA---AAAGAGAGCAGAAAGUGAAAAGACG---ACAG-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUU ...---...........(((.(((.......---...(-------((...))).((((((((((..(((((((......)))))))..))...))))))))..))).))) ( -20.50) >DroEre_CAF1 80358 91 - 1 A----------------AAAGUGAAAAGUCU---ACACGACUUUGCCGACGGCAAAGUUUUGGCUCCUUGCCAAUCGAAUUGCAAAUGGCAAUCAAAACUUAAUCAGUUU .----------------(((.(((((((((.---....))))))((((...((((....(((((.....))))).....))))...)))).............))).))) ( -24.40) >DroYak_CAF1 82373 88 - 1 AAAAA------------AAAGUGAAAAGUCG---ACAC-------CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUU .....------------(((.(((...((((---....-------.))))....((((((((((..(((((((......)))))))..))...))))))))..))).))) ( -22.30) >DroAna_CAF1 81907 87 - 1 AA---------------AAGGCGGCAAUCCGCUCCCAU-------CCGGCGGCAAAGUU-UGGCUCUCUGGAAAUCGAAUCGCAAAUUGCAAUCAAAACUUAAUCAGUUU ..---------------..(((((....)))))....(-------((((.(((......-..)).).))))).........((.....)).....(((((.....))))) ( -16.50) >consensus AAA______________AAAGUGAAAAGACG___ACAC_______CCGACGGCAAAGUUUUGGCUCUUUGCAAAUCGAAUUGCAAAUUGCAAUCAAAACUUAAUCAGUUU ..................((((........................(((..((((((........))))))...))).(((((.....)))))....))))......... (-11.27 = -11.68 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:28 2006