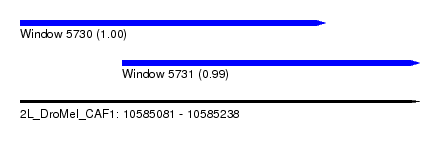
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,585,081 – 10,585,238 |
| Length | 157 |
| Max. P | 0.999432 |

| Location | 10,585,081 – 10,585,201 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.77 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
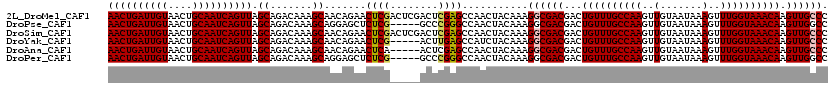
>2L_DroMel_CAF1 10585081 120 + 22407834 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAACAGAACUCGACUCGACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCC ((((((((((....)))))))))).((.......)).......(((((......)))))...........((((((...((((((((((..(.......)..)))))))))).)))))). ( -39.10) >DroPse_CAF1 75599 115 + 1 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAGGAGCUCUCG-----GCCCGGGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGGCC ((((((((((....)))))))))).((.......)).((.((.....-----)))).((((((((.....(....)...((((((((((..(.......)..)))))))))))))))))) ( -41.90) >DroSim_CAF1 73095 120 + 1 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAACAGAACUCGACUCGACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCC ((((((((((....)))))))))).((.......)).......(((((......)))))...........((((((...((((((((((..(.......)..)))))))))).)))))). ( -39.10) >DroYak_CAF1 75440 115 + 1 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAACAGAACUCG-----ACUUGAGCCAUCUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCC ((((((((((....)))))))))).((.......))...(((.(((.-----....)))...))).....((((((...((((((((((..(.......)..)))))))))).)))))). ( -35.70) >DroAna_CAF1 76324 115 + 1 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAACAGAACUCA-----ACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCC ((((((((((....))))))))))..........(((((........-----..(((.(((.........)))..))).((((((((((..(.......)..)))))))))).))))).. ( -35.70) >DroPer_CAF1 77519 115 + 1 AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAGGAGCUCUCG-----GCCCGGGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGGCC ((((((((((....)))))))))).((.......)).((.((.....-----)))).((((((((.....(....)...((((((((((..(.......)..)))))))))))))))))) ( -41.90) >consensus AACUGAUUGUAACUGCAAUCAGUUAGCAGACAAAGCAACAGAACUCG_____ACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCC ((((((((((....)))))))))).((.......)).......((((........))))...........((((((...((((((((((..(.......)..)))))))))).)))))). (-33.60 = -33.77 + 0.17)
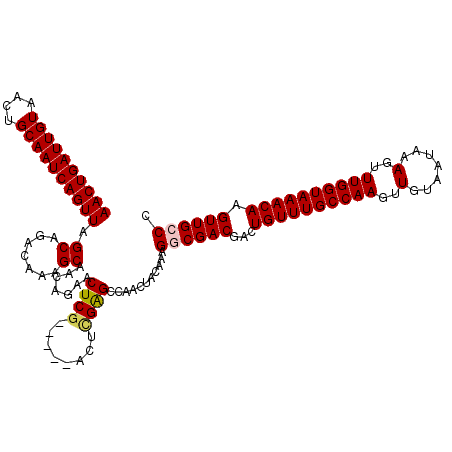
| Location | 10,585,121 – 10,585,238 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10585121 117 + 22407834 GAACUCGACUCGACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC---ACACACACUUCGGCUCCAAAUCCAACACACAAUAAG ................(((((.........((((((...((((((((((..(.......)..)))))))))).))))))..---...........))))).................... ( -27.75) >DroSec_CAF1 72619 117 + 1 GAACUCGACUCGACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC---ACACACACUUCGGCUCCAAAUCCAACACACAAUAAG ................(((((.........((((((...((((((((((..(.......)..)))))))))).))))))..---...........))))).................... ( -27.75) >DroSim_CAF1 73135 117 + 1 GAACUCGACUCGACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC---ACACACACUUCGGCUCCAAAUCCAACACACAAUAAG ................(((((.........((((((...((((((((((..(.......)..)))))))))).))))))..---...........))))).................... ( -27.75) >DroEre_CAF1 73548 112 + 1 GAACUCG-----ACUCGAGCCAUCUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC---ACACACACUUCGGCUCCAAGUCCAACACACAAUAAG ......(-----(((.(((((.........((((((...((((((((((..(.......)..)))))))))).))))))..---...........)))))..)))).............. ( -31.45) >DroYak_CAF1 75480 112 + 1 GAACUCG-----ACUUGAGCCAUCUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC---ACACACACUUCGGAUCCGACUCCAACACACAACAAG .......-----.((((.............((((((...((((((((((..(.......)..)))))))))).))))))..---...........(((......))).........)))) ( -27.00) >DroAna_CAF1 76364 111 + 1 GAACUCA-----ACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCCAACACACACACUCCUUCUCGAAC----ACACACAAUAAA .......-----..(((((...........((((((...((((((((((..(.......)..)))))))))).))))))..................)))))..----............ ( -23.85) >consensus GAACUCG_____ACUCGAGCCAACUACAAAGGCGACGACUGUUUGCCAAGUUGUAAUAAAGUUUGGUAAACAAGUUGCCCC___ACACACACUUCGGCUCCAAAUCCAACACACAAUAAG ...((((........))))...........((((((...((((((((((..(.......)..)))))))))).))))))......................................... (-24.43 = -24.48 + 0.06)

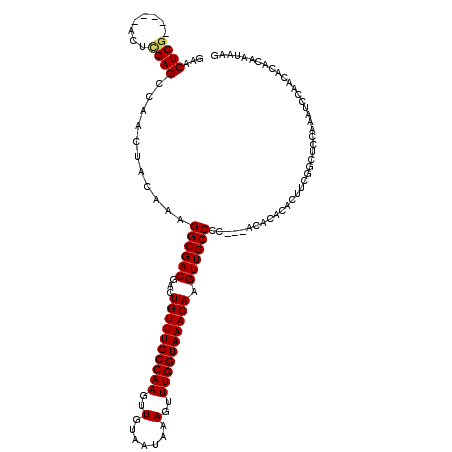
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:19 2006