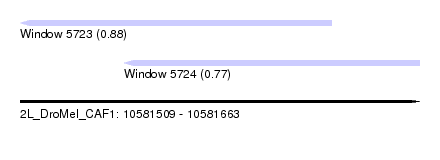
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,581,509 – 10,581,663 |
| Length | 154 |
| Max. P | 0.882524 |

| Location | 10,581,509 – 10,581,629 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
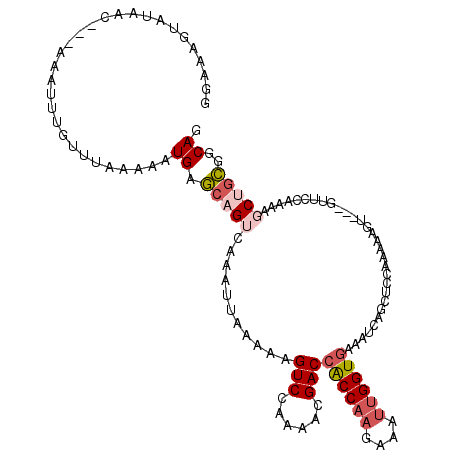
>2L_DroMel_CAF1 10581509 120 - 22407834 UCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAUCUCCGAAAGCUCAACUUCCAAAAGCUGCGGCACAGAAUCAAAUUUUUAGUUGCCAAUUCGAGGUGUUUGGUCA ............(.(((((.....((((((....))))))....((((((.(((((((...........))))..(((((((((......)))).).))))..)))))))))))))).). ( -31.00) >DroSec_CAF1 69021 117 - 1 UCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCGAAAAGU---GUUCCAAAUGCUGCGGCAGAUAAUCAAAUUUUUUGUUGCCAAUUUGAAGUGUUUGGUCA ............(.(((((((...((((((....))))))((((..((..((((((((---........(((....))).........))))))))..))..))))...)).))))).). ( -27.53) >DroSim_CAF1 69001 117 - 1 UCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCGAAAAGU---GUUCCAAAUGCUGCGGCAGAUGAUCAAAUUUUUUGUUGCCAAUUUGAAGUUUUUGGUCA ............(.(((((((...((((((....))))))((((..((..((((((((---....((..(((....)))..)).....))))))))..))..))))...).)))))).). ( -28.40) >DroEre_CAF1 69989 117 - 1 UCAAAUUAAAAAGUCAAAGACGACCACCAAGAAAUUGGUGAAAUCACCUCCAAAAAGU---GCACCAAAAUCCGCGGCAGAGAAUCAAAUUUUUAGCUGCCAAUUCGAAGUGUUCGGUCA ............(((...)))(((((((((....)))))(((..(((.((........---((..........))(((((..((........))..))))).....)).)))))))))). ( -28.10) >DroYak_CAF1 71808 117 - 1 UCAAAUUAAAAAGUCAAAAACGACCACCAAGAAAUUGGUGAAGUCAUCUCCAAAAAGU---CCACCAAAAGCUGCGGCAGAGAAUCAAAUUUUUAGUUGCCAAUUCGAAGUGUUUGGUCA .....................(((((((((....))))))..))).............---..((((((.(((..(((((..((........))..))))).......))).)))))).. ( -24.90) >DroAna_CAF1 73079 102 - 1 CCAAAUUGAUAAGUCGAAGGUGACAGCCCCGCGUCUGGUCAAAUCGGUU--AAUAAGC-------------CCAAGGCAG---AUCAAAUAUUCACCUGCCAGUUUGAGAUUUUUGGCAA ((((((((((.((.((..(((....)))...)).)).)))))(((((((--....)))-------------)...(((((---.............))))).......))).)))))... ( -24.62) >consensus UCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCAAAAAGU___GUUCCAAAAGCUGCGGCAGAGAAUCAAAUUUUUAGUUGCCAAUUCGAAGUGUUUGGUCA .....................(((((((((....)))))....................................(((((..((........))..)))))..............)))). (-15.73 = -16.48 + 0.75)


| Location | 10,581,549 – 10,581,663 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10581549 114 - 22407834 GGAAAGUAUAAG---AAAUUUCUUUAAAAAUGAGCAGUCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAUCUCCGAAAGCUCAACUUCCAAAAGCUGCGGCAC ((((.((..(((---(....))))......(((((..((..........(((......)))((((((....))))))...........))..)))))))))))....((....)).. ( -26.30) >DroSec_CAF1 69061 111 - 1 GGAAAGUAUAAC---AAAUUUCUUUAAAAAUGAGCAGUCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCGAAAAGU---GUUCCAAAUGCUGCGGCAG (((.......((---(..((((.........((((..............(((......)))((((((....))))))......)))).))))..)---)))))...(((....))). ( -21.41) >DroSim_CAF1 69041 111 - 1 GGAAAGUAUAAC---AAAUUUGUUUAAAAAUGAGCAGUCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCGAAAAGU---GUUCCAAAUGCUGCGGCAG ((((.....(((---(....)))).......((((..............(((......)))((((((....))))))......))))........---.))))...(((....))). ( -22.80) >DroEre_CAF1 70029 111 - 1 AGAAAGUAUAGC---AAAUUUGUUUGAAAAUGAGCAGUCAAAUUAAAAAGUCAAAGACGACCACCAAGAAAUUGGUGAAAUCACCUCCAAAAAGU---GCACCAAAAUCCGCGGCAG .....((...((---.((((((.(((........))).)))))).....(((......)))((((((....))))))..................---............)).)).. ( -18.50) >DroYak_CAF1 71848 111 - 1 GGAAAUUAUAAC---AAAUUUGUUUAAAAAUGAGCAGUCAAAUUAAAAAGUCAAAAACGACCACCAAGAAAUUGGUGAAGUCAUCUCCAAAAAGU---CCACCAAAAGCUGCGGCAG (((.........---....(((((((....))))))).....................(((((((((....))))))..)))...))).....((---(((.(....).)).))).. ( -20.50) >DroAna_CAF1 73116 102 - 1 ACACAGCAUAACUUUUAAUAAUUUUCAAAAUGAUGAGCCAAAUUGAUAAGUCGAAGGUGACAGCCCCGCGUCUGGUCAAAUCGGUU--AAUAAGC-------------CCAAGGCAG ....................................(((...(((((.((.((..(((....)))...)).)).)))))...((((--....)))-------------)...))).. ( -19.00) >consensus GGAAAGUAUAAC___AAAUUUGUUUAAAAAUGAGCAGUCAAAUUAAAAAGUCCAAAACGACCACCAAGAAAUUGGUGAAAUCAGCUCCAAAAAGU___GUUCCAAAAGCUGCGGCAG ..............................((.(((((...........(((......)))((((((....))))))..............................)))))..)). (-12.98 = -13.90 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:12 2006