| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,579,092 – 10,579,199 |
| Length | 107 |
| Max. P | 0.646849 |

| Location | 10,579,092 – 10,579,199 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -18.01 |
| Energy contribution | -19.40 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
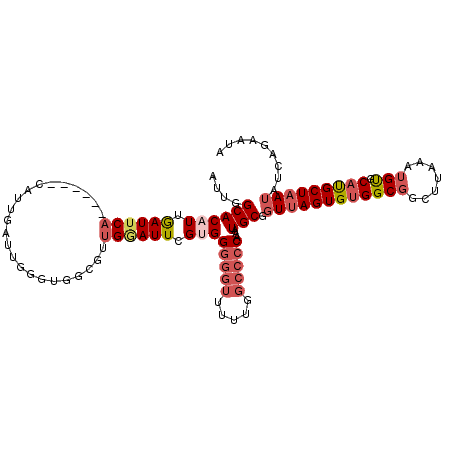
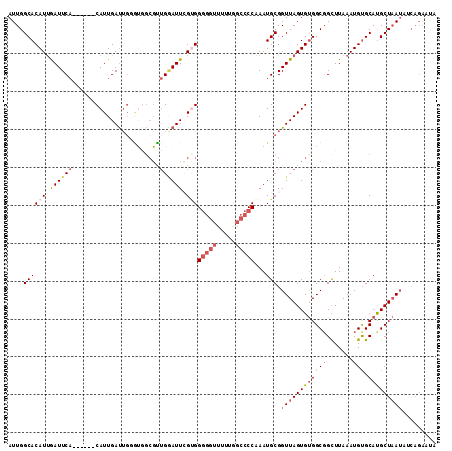
>2L_DroMel_CAF1 10579092 107 - 22407834 AUUGGCACAUUGAUUCA------CAUUGAUUGUAUGGCGUUGGAUUGGUGGGGGUUUUUAGCCCCAAAUGCGGUUAGUGUGGCGGCUUAAAUGUGCACGCUAAUAUCACGUUC ....(((((((...(((------((((((((((((..((......))...(((((.....)))))..))))))))))))))).......)))))))................. ( -32.40) >DroSec_CAF1 66632 113 - 1 AUUGGCACAUUGAUUCACAUUCACAUUGAUUGGGUGGCGUUGGAUUCGUGGGGGUUUUUGGCCCCAAAUGCGGUUAGUGUGGCGGCUUAAAUGUGCAUGCUAAUAUCAGAAUC ....(((((((.........((((((((((((.((.(((.......))).(((((.....)))))..)).)))))))))))).......)))))))................. ( -33.19) >DroSim_CAF1 66616 113 - 1 AUUGGCACAUUGAUUCACAUUCACAUUGAUUGGGUGGCGCUGGAUUCGUGGGGGUUUUUGGCCCCAAAUGCGGUUAGUGUGGCGGUUUAAAUGUGCAUGCUAAUAUCAGAAUC ....(((((((.....((..((((((((((((.((..(((.......)))(((((.....)))))..)).))))))))))))..))...)))))))................. ( -34.20) >DroEre_CAF1 67611 106 - 1 AUUGGCACAUUGAUUCA------CAUUGAUUUUGUAGCGCUGCAUUCGAGGGGGUUUU-UGCCCCAAAUGCGGUUAGUGCGGCUGCUUAAAUGUGCAUGCUCAUCGCUGAGCA ....(((((((.....(------((.......)))(((((((((((((..(((((...-.))))).....)))...))))))).)))..))))))).((((((....)))))) ( -39.50) >DroYak_CAF1 69431 106 - 1 AUUGGCACAUUGAUUCA------CAUUGAUUGGGCGGCGUUGAAUUCGCGGGGGUUUU-GGCCCCAAAUGCGGUUAGUGUGGCGGCUUAAAUGUGCAUGCUAAGCGCAGAGCA ..((.(((((((((.(.------((((......((((........)))).(((((...-.))))).)))).)))))))))).))((((...(((((.......))))))))). ( -36.20) >DroAna_CAF1 70659 91 - 1 AUUGGCACAUCAAUUCC------CAUUGACGG---------GGAUUGGUGG-------UGCCACCGAAUGCGGUUAGUGUGGCGGCUUAAAUGCGCACGCUAAUCAGAGACUA ..(((((((((((((((------(......))---------)))))))).)-------)))))........(((((((((((((.......))).))))))))))........ ( -40.80) >consensus AUUGGCACAUUGAUUCA______CAUUGAUUGGGUGGCGUUGGAUUCGUGGGGGUUUUUGGCCCCAAAUGCGGUUAGUGUGGCGGCUUAAAUGUGCAUGCUAAUAUCAGAAUA ....((((((.((((((.......................)))))).)))(((((.....)))))...))).((((((((((((.......))).)))))))))......... (-18.01 = -19.40 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:11 2006