| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,555,795 – 10,555,914 |
| Length | 119 |
| Max. P | 0.640644 |

| Location | 10,555,795 – 10,555,914 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -19.60 |
| Energy contribution | -18.52 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
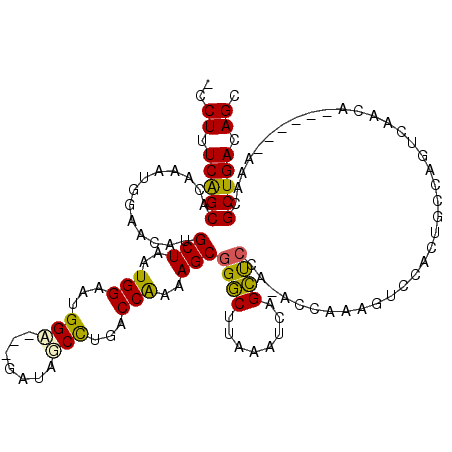
>2L_DroMel_CAF1 10555795 119 - 22407834 -CCUUUCGGCACAAAUGUAACAUGCUAAUGGAGUGGAGAAGAUAUCCUGACCAAAAGCGGGCUUAAAUCAGCUCCAAACCAAAGUCCACUGCCAGUGGACGGCGGCCAAAUGCUGACAGC -.((.(((((((....))....((((..(((..(((((..(((..((((........)))).....)))..)))))..)))..((((((.....)))))))))).......))))).)). ( -36.20) >DroPse_CAF1 46244 110 - 1 GUCUUUCAGCACAAAUGGAACAUGCUAAUGGAACGGA---GAUGGCUCGACCGUAAGCGGACUUAAGUCCGUCCCG-ACCAACGUCCAGAGACGGUCAACA------AAACGCUGACAGC ..((.(((((......((.((..(((.((((....((---(....)))..)))).)))((((....)))))).))(-(((...(((....)))))))....------....))))).)). ( -33.30) >DroEre_CAF1 44216 115 - 1 -CCUUUCGGCACAAAUGAAACAUGCUAAUGGAGUGGA---GUUAUCCUGACCAAAAGCGGGCUUAACUCAGCUCCA-ACCAAAGUCCACUGCCAGUGGACAGCGGCCAAACGCUGACAGC -.((.(((((.............(((..((((((.((---((((.((((........))))..)))))).))))))-......((((((.....)))))))))(......)))))).)). ( -41.30) >DroYak_CAF1 45634 116 - 1 -CCUUUCAGCACAAAUGAAACAUGCUAAUGGAGUGGA---GAUAUCCUAACCAAAAGCGGGCUCAAAUCAGCUCCAAACCAAAGUCCACUGCCAGUGGACAGCGGCCAAAUGCUGACAGC -.((.((((((............(((..(((...(((---....)))...)))..)))(((((......))).))...((...((((((.....))))))...)).....)))))).)). ( -33.70) >DroAna_CAF1 47859 109 - 1 -UCUUUCGGCACAAAUGGAACAUGCUAAUGGAAUGGC---GAUGGCUUGGCCGAAAGCUGGCUUAACUCAGCUCCU-ACCAACGUCCAGGGGCAGUCAACA------AAACGCUGACAGC -.((.(((((.......((...((((..((((.(((.---..((((...))))..((((((......))))))...-.)))...))))..)))).))....------....))))).)). ( -30.86) >DroPer_CAF1 46228 110 - 1 GUCUUUCAGCACAAAUGGAACAUGCUAAUGGAACGGA---GAUGGCUCGACCGUAAGCGGACUUAAGUCCGUCCCG-ACCAACGUCCAGAGACGGUCAACA------AAACGCUGACAGC ..((.(((((......((.((..(((.((((....((---(....)))..)))).)))((((....)))))).))(-(((...(((....)))))))....------....))))).)). ( -33.30) >consensus _CCUUUCAGCACAAAUGGAACAUGCUAAUGGAAUGGA___GAUAGCCUGACCAAAAGCGGGCUUAAAUCAGCUCCA_ACCAAAGUCCACUGCCAGUCAACA______AAACGCUGACAGC ..((.(((((.............(((..(((...(((.......)))...)))..)))((((........)))).....................................))))).)). (-19.60 = -18.52 + -1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:03 2006