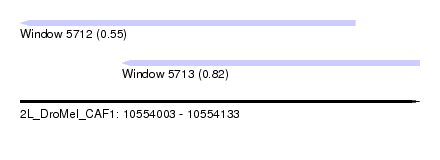
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,554,003 – 10,554,133 |
| Length | 130 |
| Max. P | 0.823538 |

| Location | 10,554,003 – 10,554,112 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -24.11 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10554003 109 - 22407834 -AUUGGC---CCACAUUCACAUGCCACACACGCAGCCGGCGAAGAAAUUUAAAACGUCAACAAGACCGUUGACAGUUGGCGCUGUGCAGGACUCGUAAAAAA-----CUGCG--UCACUC -..((((---............)))).(((.((.(((((((((....))).....((((((......)))))).)))))))))))((((.............-----)))).--...... ( -29.62) >DroPse_CAF1 44578 118 - 1 GACCGACCUUUCGCGUUCGCAUGCCACACACGCAGCCUGCAAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGCGCUGUGCAGGAAUCGUAAAAAAAAACCCCCCG--CCACAU ...((((((...((....))......((((.((.(((.(((((....))).....((((((......)))))).)).))))))))).)))..))).................--...... ( -25.60) >DroEre_CAF1 42410 109 - 1 -AUUGGC---CCACAUUCACGUGCCACACACGCAGCCAGCGGAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGCGCUGUGCAGGACUCGUAAAAAA-----CCGCG--CCACUC -..((((---(.......(((.(((.((((.((.(((((((((....))).....((((((......)))))).))))))))))))..)).).)))......-----..).)--)))... ( -33.66) >DroYak_CAF1 43730 109 - 1 -AGUGGC---CCACAUUCACGUGCCACACACGCAGCCAGCGAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGCGCUGUGCAGGACUCGUAAAAAA-----CUGCG--CCACUC -((((((---.((.....(((.(((.((((.((.(((((((((....))).....((((((......)))))).))))))))))))..)).).)))......-----.)).)--))))). ( -38.90) >DroAna_CAF1 46478 111 - 1 -ACAACC---UCGCACUCACAUGCCGCACACGCAGCUAGCAAAGAAAUUUAAAACGUCAACAAGCCCAUUGACAGUUGGCGCUGUGCAGGACUCGUAAAAAA-----AGGCUGCCCCCUC -....((---(.(((......))).(((((.((.(((((((((....))).....(((((........))))).))))))))))))))))............-----(((......))). ( -27.10) >DroPer_CAF1 44563 118 - 1 GACCGACCUUUCGCGUUCGCAUGCCACACACGCAGCCUGCAAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGCGCUGUGCAGGAAUCGUAAAAAAAAACCCCCCG--CCACAU ...((((((...((....))......((((.((.(((.(((((....))).....((((((......)))))).)).))))))))).)))..))).................--...... ( -25.60) >consensus _ACUGGC___CCACAUUCACAUGCCACACACGCAGCCAGCAAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGCGCUGUGCAGGACUCGUAAAAAA_____CCGCG__CCACUC ..................((...((.((((.((.(((((((((....))).....((((((......)))))).))))))))))))..))....))........................ (-24.11 = -23.78 + -0.33)

| Location | 10,554,036 – 10,554,133 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10554036 97 - 22407834 CCA-CUC------CAUGCCCCCA------------ACCCA-AUUGGCCCACAUUCACAUGCCACACACGCAGCCGGCGAAGAAAUUUAAAACGUCAACAAGACCGUUGACAGUUGGC ...-...------..........------------.....-..((((............))))........(((((((((....))).....((((((......)))))).)))))) ( -19.80) >DroSec_CAF1 41818 98 - 1 CCA-CUC------CAUGCCCCCU------------CCCAAAAUUGGCCCACAUUCACAUGCCACACACGCAGUCGGCGAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGC (((-.((------..((((..((------------........((((............)))).......))..))))..))..........((((((......))))))...))). ( -18.16) >DroSim_CAF1 41464 98 - 1 CCA-CUC------CAUGCCCCCU------------CCCAAACUUGGCCCACAUUCACAUGCCACACACGCAGCCGGCGAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGC ...-...------..........------------........((((............))))........(((((((((....))).....((((((......)))))).)))))) ( -19.80) >DroEre_CAF1 42443 94 - 1 CCA-CUC------CAUUCCCGCU------------CC----AUUGGCCCACAUUCACGUGCCACACACGCAGCCAGCGGAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGC ...-..(------((......((------------((----.(((((.........((((.....))))..))))).))))...........((((((......))))))...))). ( -23.50) >DroYak_CAF1 43763 113 - 1 CCAUCUCCAUCCCCAUUCCCAUUCCAAGCCUGCUCCC----AGUGGCCCACAUUCACGUGCCACACACGCAGCCAGCGAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGC ..............................(((....----.(((((.(........).)))))....)))(((((((((....))).....((((((......)))))).)))))) ( -26.90) >DroAna_CAF1 46513 84 - 1 CCG-CAC----------------------------CC----ACAACCUCGCACUCACAUGCCGCACACGCAGCUAGCAAAGAAAUUUAAAACGUCAACAAGCCCAUUGACAGUUGGC ...-...----------------------------..----........(((......))).((....)).(((((((((....))).....(((((........))))).)))))) ( -13.60) >consensus CCA_CUC______CAUGCCCCCU____________CC____AUUGGCCCACAUUCACAUGCCACACACGCAGCCAGCGAAGAAAUUUAAAACGUCAACAAGCCCGUUGACAGUUGGC ...........................................((((............))))........(((((((((....))).....((((((......)))))).)))))) (-17.37 = -17.23 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:02 2006