
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,553,546 – 10,553,642 |
| Length | 96 |
| Max. P | 0.986683 |

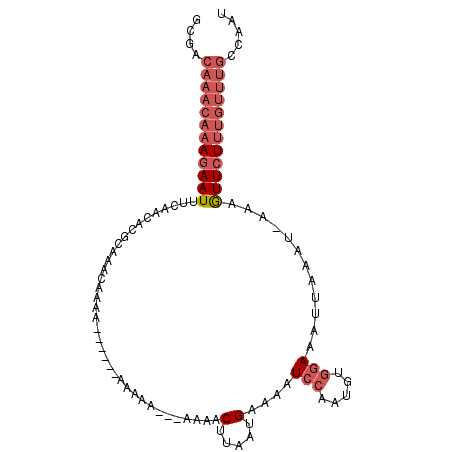
| Location | 10,553,546 – 10,553,642 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -16.08 |
| Consensus MFE | -7.79 |
| Energy contribution | -9.32 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
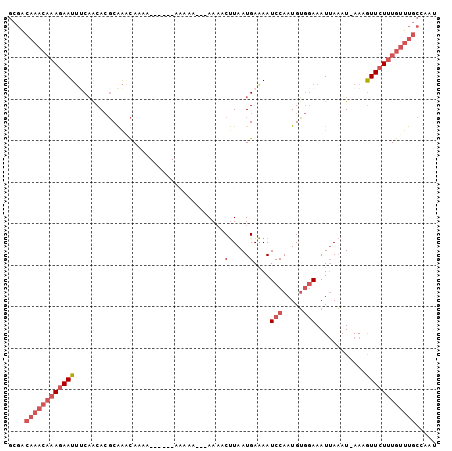
>2L_DroMel_CAF1 10553546 96 + 22407834 GCGACAAACAAAGAAUUUCAGCACGCAAACAAAA------AAAAAAAAAAAACUUAAUGAAAAUCCAAUGUGGAAAUUAAAU-AAUGUUCUUUGUUUGCCAAU ..(.((((((((((((((((..............------.................))))).(((.....)))........-....))))))))))).)... ( -15.48) >DroPse_CAF1 44047 92 + 1 G----------AAAAUUCCCACAGCCGAAUAAAAUGUUUAAAGCGAAAAAUCCCUAAUGAGAAUCCAAUGG-AAAAUUACAUCAAAAUUCUUUGUUUAAACAC .----------...((((........))))....((((((((.((((...((......))((((...(((.-.......)))....)))))))))))))))). ( -7.60) >DroSec_CAF1 41335 93 + 1 GCGACAAACAAAGAAUUUCAGCACGCAAACACAA------AAAAA---AAAACUUAAUGAAAAUCCAAUGUGGAAAUUAAAU-AAUGUUCUUUGUUUGCCAAU ..(.((((((((((((((((..............------.....---.........))))).(((.....)))........-....))))))))))).)... ( -15.59) >DroSim_CAF1 40982 96 + 1 GCGACAAACAAAGAAUUUCAACACGCAAACAAAA------AAAAAAGAAAAACUUAAUGAAAAUCCAAUGUGGAAAUUAAAU-AAUGUUCUUUGUUUGCCAAU ..(.((((((((((((..................------....(((.....)))........(((.....)))........-...)))))))))))).)... ( -15.10) >DroEre_CAF1 42003 85 + 1 GCGACAAACAAAGAAUUUCAGCACGCAACCA-----------------AAAACUUAAUGAAAAUCCAAUGUGGAAAUUAAAU-AAAGUUCUUUGUUUGCGAAU .((.((((((((((((((..(....).....-----------------...............(((.....)))........-.))))))))))))))))... ( -18.10) >DroYak_CAF1 43313 85 + 1 GCGGCAAACAAAGAAUGGCAACACGCAGCCA-----------------AAAACUUAAUGAAAAUCCAAUGUGGAAAUGAAAU-AAAGUUCUUUGUUUGCGAAU ...((((((((((((((((........))).-----------------...............(((.....)))........-...))))))))))))).... ( -24.60) >consensus GCGACAAACAAAGAAUUUCAACACGCAAACAAAA______AAAAA___AAAACUUAAUGAAAAUCCAAUGUGGAAAUUAAAU_AAAGUUCUUUGUUUGCCAAU ....((((((((((((....................................(.....)....(((.....)))............))))))))))))..... ( -7.79 = -9.32 + 1.53)

| Location | 10,553,546 – 10,553,642 |
|---|---|
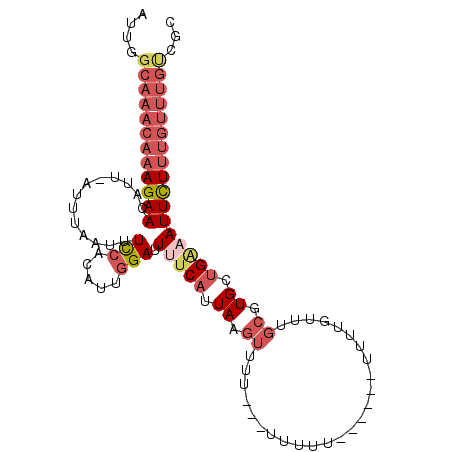
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -8.96 |
| Energy contribution | -10.96 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
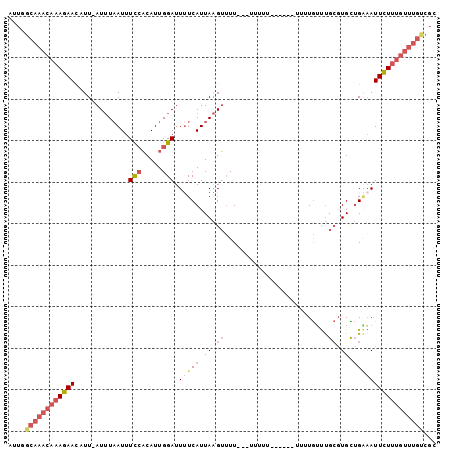
>2L_DroMel_CAF1 10553546 96 - 22407834 AUUGGCAAACAAAGAACAUU-AUUUAAUUUCCACAUUGGAUUUUCAUUAAGUUUUUUUUUUUU------UUUUGUUUGCGUGCUGAAAUUCUUUGUUUGUCGC ...(((((((((((((....-(((((((.(((.....))).....)))))))..........(------(((.((......)).))))))))))))))))).. ( -20.20) >DroPse_CAF1 44047 92 - 1 GUGUUUAAACAAAGAAUUUUGAUGUAAUUUU-CCAUUGGAUUCUCAUUAGGGAUUUUUCGCUUUAAACAUUUUAUUCGGCUGUGGGAAUUUU----------C ((((((((((.(((((((((((((......(-((...)))....))))).)))))))).).)))))))))...((((........))))...----------. ( -13.80) >DroSec_CAF1 41335 93 - 1 AUUGGCAAACAAAGAACAUU-AUUUAAUUUCCACAUUGGAUUUUCAUUAAGUUUU---UUUUU------UUGUGUUUGCGUGCUGAAAUUCUUUGUUUGUCGC ...(((((((((((((....-(((((((.(((.....))).....)))))))...---..(((------(.(..(....)..).))))))))))))))))).. ( -20.30) >DroSim_CAF1 40982 96 - 1 AUUGGCAAACAAAGAACAUU-AUUUAAUUUCCACAUUGGAUUUUCAUUAAGUUUUUCUUUUUU------UUUUGUUUGCGUGUUGAAAUUCUUUGUUUGUCGC ...(((((((((((((....-........(((.....))).(((((....((...........------........))....)))))))))))))))))).. ( -20.51) >DroEre_CAF1 42003 85 - 1 AUUCGCAAACAAAGAACUUU-AUUUAAUUUCCACAUUGGAUUUUCAUUAAGUUUU-----------------UGGUUGCGUGCUGAAAUUCUUUGUUUGUCGC ...(((((((((((((....-(((((((.(((.....))).....)))))))(((-----------------..((.....))..)))))))))))))).)). ( -20.10) >DroYak_CAF1 43313 85 - 1 AUUCGCAAACAAAGAACUUU-AUUUCAUUUCCACAUUGGAUUUUCAUUAAGUUUU-----------------UGGCUGCGUGUUGCCAUUCUUUGUUUGCCGC ....(((((((((((((((.-........(((.....)))........))))...-----------------((((........)))).)))))))))))... ( -23.83) >consensus AUUGGCAAACAAAGAACAUU_AUUUAAUUUCCACAUUGGAUUUUCAUUAAGUUUU___UUUUU______UUUUGUUUGCGUGCUGAAAUUCUUUGUUUGUCGC ....((((((((((((.............(((.....))).(((((.((.((.........................)).)).)))))))))))))))))... ( -8.96 = -10.96 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:00 2006