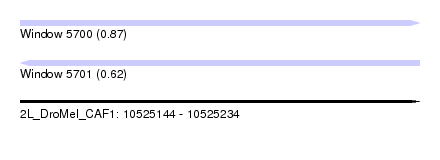
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,525,144 – 10,525,234 |
| Length | 90 |
| Max. P | 0.867433 |

| Location | 10,525,144 – 10,525,234 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
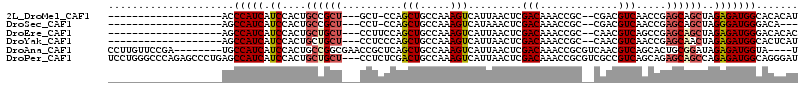
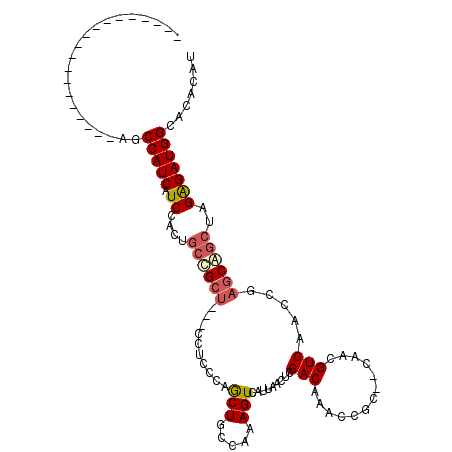
>2L_DroMel_CAF1 10525144 90 + 22407834 AUGUGUGCCAUCUCUAGCUGCUCGGUUGACGUCG--GCGGUUUGUCGAGUUAAUGACUUUGGCAGCUGG-AGC---AGCGGCAGUGGAUGAUGGGU------------------- .....((((.((((((((((((.(((((((.(((--((.....))))))))...))))..)))))))))-)).---)..)))).............------------------- ( -33.10) >DroSec_CAF1 13495 87 + 1 ---UGUCCCAUCCCUAGCUGCUCGGUUGACGUCG--GCGGUUUGUCGAGUUUAUGACUUUGGCAGCUGG-AGG---AGCGGCAGUGGAUGAUGGCU------------------- ---.(((.(((((((.(((((((.((.(((.(((--((.....)))))))).))..(((..(...)..)-)))---)))))))).)))))..))).------------------- ( -33.10) >DroEre_CAF1 13832 91 + 1 GUGUGUCCCAUCUCUAGCUGCUCGGCUGACGUUG--GCGGUUUGUCGAGUUAAUGACUUUGGCAGCUGGAAGG---AGCAGCAGUGGAUGAUGGCU------------------- .......((((((((((((((((..((..(((((--.(((...((((......)))).))).)))).)..)))---))))))..)))).)))))..------------------- ( -32.00) >DroYak_CAF1 13923 91 + 1 AUGAGUGCCAUCUCUAGUUGCUCGGUUGACGUUG--GCGGUUUGUCGAGUUAAUGACUUUGGCAGCUGGGAGG---AGCAGCAGUGGAUGAUGGCU------------------- ......(((((((((((((((((..((.(.((((--.(((...((((......)))).))).))))).))..)---))))))..)))).)))))).------------------- ( -33.40) >DroAna_CAF1 13813 103 + 1 A----UACCAUCUCUAUCCGCAGUGCUGACGUUGACGCGGUUUGUCGAGUUAAUGACUUUGGCAGCUGAGCGGUUCGCCGGCAGUGGAUGAUGGCA--------UCGGAACAAGG .----..(((((...((((((..(((..((((((((.(((....))).)))))))...)..)))((((.(((...))))))).)))))))))))..--------........... ( -35.30) >DroPer_CAF1 14844 112 + 1 AUCCCUGCCAUCUCUGGCUGCUCUGCUGACGGCGACGCGGUUUGUCGAGUUAAUGACUUUGGCAGUCGAGAGG---AGCAGCAGUGGAUGAUGGCUCAGGGCUCUGGGCCCAGGA ...((((((((((((.((((((((.(((((..(((((.....))))).(((((.....))))).))).)).))---))))))...))).))))))...((((.....))))))). ( -50.20) >consensus AUGUGUGCCAUCUCUAGCUGCUCGGCUGACGUCG__GCGGUUUGUCGAGUUAAUGACUUUGGCAGCUGGGAGG___AGCAGCAGUGGAUGAUGGCU___________________ .......(((((((((((((((((((((.((......))....((((......)))).....))))))........))))))..)))).)))))..................... (-22.77 = -22.88 + 0.11)

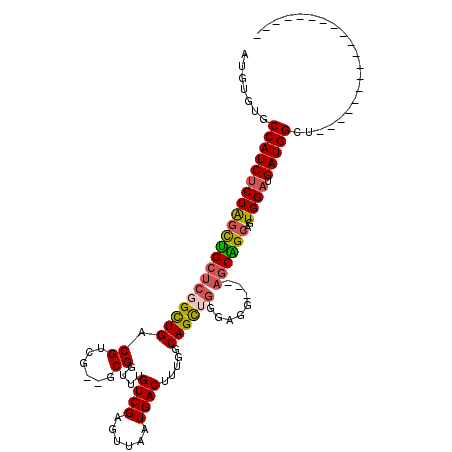
| Location | 10,525,144 – 10,525,234 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10525144 90 - 22407834 -------------------ACCCAUCAUCCACUGCCGCU---GCU-CCAGCUGCCAAAGUCAUUAACUCGACAAACCGC--CGACGUCAACCGAGCAGCUAGAGAUGGCACACAU -------------------.............(((((..---.((-(.((((((....(((........))).......--((........)).)))))).))).)))))..... ( -23.00) >DroSec_CAF1 13495 87 - 1 -------------------AGCCAUCAUCCACUGCCGCU---CCU-CCAGCUGCCAAAGUCAUAAACUCGACAAACCGC--CGACGUCAACCGAGCAGCUAGGGAUGGGACA--- -------------------...............(((..---(((-..((((((....(((........))).......--((........)).)))))))))..)))....--- ( -20.30) >DroEre_CAF1 13832 91 - 1 -------------------AGCCAUCAUCCACUGCUGCU---CCUUCCAGCUGCCAAAGUCAUUAACUCGACAAACCGC--CAACGUCAGCCGAGCAGCUAGAGAUGGGACACAC -------------------..(((((.((....((((((---(......((((.....(((........)))....((.--...)).)))).)))))))..)))))))....... ( -24.30) >DroYak_CAF1 13923 91 - 1 -------------------AGCCAUCAUCCACUGCUGCU---CCUCCCAGCUGCCAAAGUCAUUAACUCGACAAACCGC--CAACGUCAACCGAGCAACUAGAGAUGGCACUCAU -------------------.((((((.((....(.((((---(......(((.....))).........(((.......--....)))....))))).)..))))))))...... ( -19.80) >DroAna_CAF1 13813 103 - 1 CCUUGUUCCGA--------UGCCAUCAUCCACUGCCGGCGAACCGCUCAGCUGCCAAAGUCAUUAACUCGACAAACCGCGUCAACGUCAGCACUGCGGAUAGAGAUGGUA----U ..........(--------(((((((..((......))....((((...((((.....(((........))).....(((....)))))))...)))).....)))))))----) ( -27.00) >DroPer_CAF1 14844 112 - 1 UCCUGGGCCCAGAGCCCUGAGCCAUCAUCCACUGCUGCU---CCUCUCGACUGCCAAAGUCAUUAACUCGACAAACCGCGUCGCCGUCAGCAGAGCAGCCAGAGAUGGCAGGGAU ((((((((.....))))...((((((.((....((((((---(..((.(((.((...(((.....))).(((.......))))).)))))..)))))))..)))))))).)))). ( -42.20) >consensus ___________________AGCCAUCAUCCACUGCCGCU___CCUCCCAGCUGCCAAAGUCAUUAACUCGACAAACCGC__CAACGUCAACCGAGCAGCUAGAGAUGGCACACAU .....................(((((.((....((((((..........(((.....))).........(((.............))).....))))))..)))))))....... (-13.65 = -14.20 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:50 2006