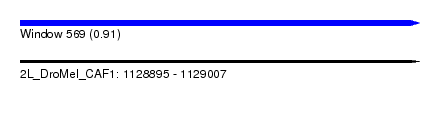
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,128,895 – 1,129,007 |
| Length | 112 |
| Max. P | 0.906789 |

| Location | 1,128,895 – 1,129,007 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -17.93 |
| Energy contribution | -20.43 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1128895 112 + 22407834 AAUUCAGGCAAAACAAAACUUUGCUUGCAAAAUUGCUGGCAACUAGAGGUGGUCAGCUGUCGAUUAUCGUAGUUGGCAACACUAAUCGAUAGG---ACAGAAAGCGUUAUGCAUU .....(((((((.......)))))))(((.....((((((.((.....)).))))))((((..((((((((((.(....)))))..)))))))---)))..........)))... ( -29.90) >DroSec_CAF1 15476 104 + 1 --------UAAAACAAAACUUUGCUUGCAAAAGUGCUGGCAAUUGGAGGUGGCCCGCUGUCGAUUAUUGUAGUUGGCAACAGUAAUCGAUAGG---AUAGAAAGCGUUAUGCAUU --------.................((((.((.(((((((.((.....)).)))..((((((((((((((.(....).)))))))))))))).---......)))))).)))).. ( -28.60) >DroSim_CAF1 15341 112 + 1 AAUUCCUGCAAAACAAAACUUUGCUUGCAAAAUUGCUGGCAACUGGAGGUGGCCCGCUGUCGAUUAUUGUAGUUGGCAACACUAAUCGAUAGG---ACAGAAAGCGUUAUGCAUU ......((((.(((......(((((.(((....))).)))))(((..((....)).((((((((((.(((.(....).))).)))))))))).---.))).....))).)))).. ( -32.60) >DroEre_CAF1 15868 110 + 1 AAUUUCUGCAAAACAAAACUUUGCUUGCAA-AUUGCUGGCAACUAGAGGUGGUAAGCUGCCGAUUUUUGUAGUUGGCAACACCAAUCGAUAG----ACAAAAAACGUUAUGCAUU ......((((.(((......(((((.((..-...)).)))))(((..((((((..(.((((((((.....)))))))).)))).)))..)))----.........))).)))).. ( -30.00) >DroYak_CAF1 16262 111 + 1 AAUUUCUGCAAAACAAAACUUUGCUUGCAA-AUUGCUGGCAACUAGAGGUGGUCACCUGUCGAUUGUUGUAGUUGGCAACACUAAUCGAUAGG---ACAAAAACCGUUAUGCCUU .......(((.(((......(((((.((..-...)).))))).....(((.....(((((((((((((((.....))))))...)))))))))---......)))))).)))... ( -33.70) >DroAna_CAF1 16206 94 + 1 -------UUAAAACAAAACUUUGCAUGCAA-AUUGCUGGCAACUAGAGAUGGAAGAGUGUUGUUGCAAGCAGUUGGCAACACUAAUCGAUAAGUGGAUACUA------------- -------..................(((.(-((((((.(((((..((.((......)).))))))).))))))).))).((((........)))).......------------- ( -21.30) >consensus AAUU_CUGCAAAACAAAACUUUGCUUGCAA_AUUGCUGGCAACUAGAGGUGGUCAGCUGUCGAUUAUUGUAGUUGGCAACACUAAUCGAUAGG___ACAGAAAGCGUUAUGCAUU .......(((.(((......(((((.(((....))).)))))..............((((((((((.(((.(....).))).)))))))))).............))).)))... (-17.93 = -20.43 + 2.50)

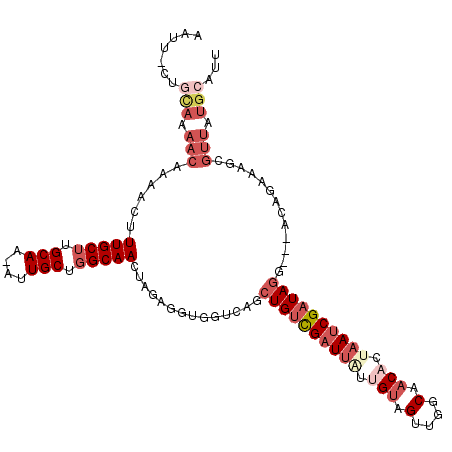
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:55 2006