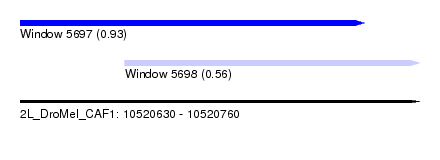
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,520,630 – 10,520,760 |
| Length | 130 |
| Max. P | 0.930730 |

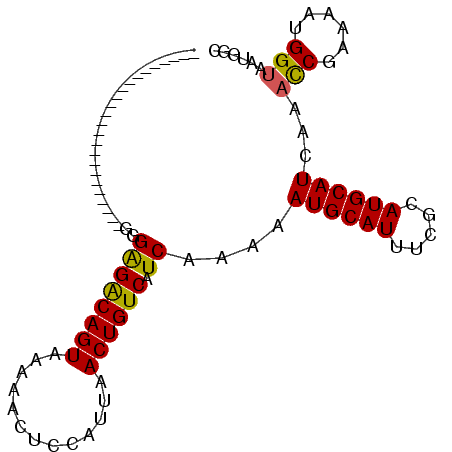
| Location | 10,520,630 – 10,520,742 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.78 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.15 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10520630 112 + 22407834 GCCCCCCCAAAAAAAACCCUACUUCCACAUAAAGUCCACGUUGGUGGGGAGGGGGA--GUGGGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA .(((((((............((((.......))))((((....)))))).))))).--.(((((((((.............)))))).)))...((((((.....))))))... ( -30.02) >DroSec_CAF1 8936 97 + 1 GCC---CCAAAAAUACCCCCACUGCCACAUAAAG------------AGGAAGGGGG--UGGAGGCAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA ...---.......(((((((....((.(.....)------------.))..)))))--))((((((((.............)))))).))....((((((.....))))))... ( -24.62) >DroSim_CAF1 8947 99 + 1 GCC---CCAAAAAUACCCACACUGCCACAUAACA------------AGGAUGCGGGAUGGGGGGCAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA .((---(((......(((.((...((........------------.)).)).))).)))))((((((.............)))))).......((((((.....))))))... ( -23.32) >DroEre_CAF1 9354 95 + 1 CCC---CCAAAAAAAUCUCCACUGCCACAUAAACUCCACGU----------------UGGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA ..(---((((..............................)----------------)))).((((((.............)))))).......((((((.....))))))... ( -14.43) >DroYak_CAF1 9260 95 + 1 CCC---CCAAAAAUACUCCCACUGCCACAUAAAGUCCACGU----------------UGGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA ...---..........(((((.((..((.....))...)).----------------)))))((((((.............)))))).......((((((.....))))))... ( -17.12) >DroPer_CAF1 9529 90 + 1 CCC---CCCAG-----------CCUCCCAUAAAGUCCACGCUC--------AGAGA--CAGAGACAGUAAAAACUCCAUUAACCGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA ...---.....-----------...........((((......--------.).))--).(((((.((.............)).))).))....((((((.....))))))... ( -8.52) >consensus CCC___CCAAAAAUACCCCCACUGCCACAUAAAGUCCACGU__________GGGG___GGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAA ............................................................((((((((.............)))))).))....((((((.....))))))... (-10.43 = -10.15 + -0.28)

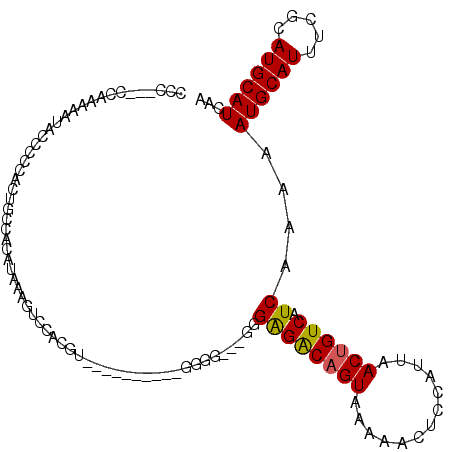
| Location | 10,520,664 – 10,520,760 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -12.13 |
| Energy contribution | -11.69 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10520664 96 + 22407834 UCCACGUUGGUGGGGAGGGGGA--GUGGGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAAUGGUAAUGGC .(((..(((((..(((.((..(--((((...(((....))))))))..)).)).......((((((.....)))))))..)))))...)))....... ( -23.10) >DroSec_CAF1 8967 84 + 1 ------------AGGAAGGGGG--UGGAGGCAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAAUGGUAAUGGC ------------....((..((--(((((..........)))))))..))(((((.....((((((.....))))))...(((......))).))))) ( -19.60) >DroSim_CAF1 8978 86 + 1 ------------AGGAUGCGGGAUGGGGGGCAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAAUGGUAAUGGC ------------.(.(((((..(((.(.((((((.............))))))........).)))..))))).).....(((......)))...... ( -21.52) >DroEre_CAF1 9385 82 + 1 UCCACGU----------------UGGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAACGGUAAUGGC .(((..(----------------(((..((((((.............)))))).))))..((((((.....))))))...((((....))))..))). ( -17.22) >DroYak_CAF1 9291 82 + 1 UCCACGU----------------UGGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAAUGGUAAUGGC .(((..(----------------(((((((((((.............)))))).))....((((((.....))))))....))))...)))....... ( -18.02) >DroAna_CAF1 9534 73 + 1 -------------------------UGAGGCAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAAUCCGAAAUGGCCAUGGC -------------------------(((((((((.............)))))).)))......(((((((.((.......)).)))))))((....)) ( -14.72) >consensus ________________________GGGAGACAGUAAAAACUCCAUUAACUGUCAUCAAAAAUGCAUUUCGCAUGCAUCAAACCGAAAAUGGUAAUGGC ..........................((((((((.............)))))).))....((((((.....))))))...(((......)))...... (-12.13 = -11.69 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:47 2006