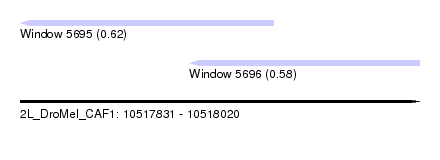
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,517,831 – 10,518,020 |
| Length | 189 |
| Max. P | 0.618053 |

| Location | 10,517,831 – 10,517,951 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
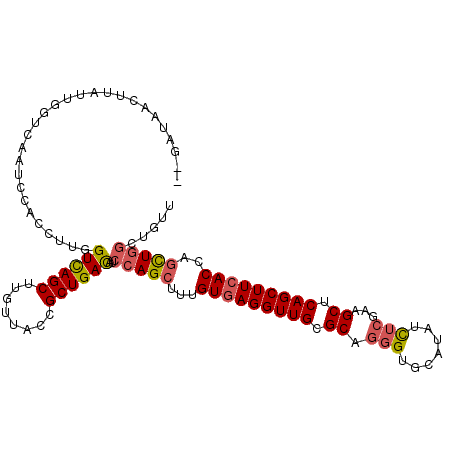
>2L_DroMel_CAF1 10517831 120 - 22407834 GGGUGCAUAUCUCGAAGCUCAGCUUCACUAGCUGGCUGUUGUUGAUGGUCAGCGAUUUGAGGGUGCGCGGAAAUCCCUUGAUGGGAUACUCGCUCCGCUUGCUGAUUACCAUGCGCACCU ((((((.........(((.(((((.....))))))))((...((.((((((((((.....(((.(((.(...(((((.....))))).).))))))..)))))))))).)).)))))))) ( -48.30) >DroVir_CAF1 6960 120 - 1 GCGUACAAAUCUCAAAGCUGAGCUUCAUCAAUUGGAUAUUGUUGAUGGUCAGCGAUUUGAGAGUGCGCGGAAAACCUUUAACUGGAUAUUCGCUGCGUUUAUUGAUGGUCAUGCGCAGCU ((((((...((((((.(((((.(.(((.((((.....)))).))).).)))))...))))))))))))(((...((.......))...)))(((((((....((.....)).))))))). ( -44.00) >DroGri_CAF1 6781 120 - 1 GCGUGCAAAUCUCAAAGCUGAGCUUCAUCAACUGUAUAUUGUUGAUGCUCAGAGAUUUGAGAGUUCGCGGAAAACCUUUAAUGGGGUAUUCGCUGCGCUUGUUGAUUGACAUACGCUGCU .((((.((.(((((((.((((((...((((((........))))))))))))...))))))).))))))(((.(((((....))))).)))((.(((..((((....))))..))).)). ( -45.50) >DroMoj_CAF1 8495 120 - 1 GCGUGCAAAUCUCGAAGCUCAGCUUCACCAAUUGAAUAUUUUUGAUGGUCAGCGAUUUGAGGGUGCGCGGAAAACCUUUGAUGGGAUACUCGCUACGCUUGUUGAUUGUCAUGCGCAGCU (((((((......(((((...)))))................(((..((((((.....(((.(((.((((....(((.....)))....))))))).)))))))))..)))))))).)). ( -37.00) >DroAna_CAF1 6302 120 - 1 GCGAGCAUAUCUCGAAACUGAGCUUCACCAGUUGACUGUUGUUGAUUGUUAACGACUUCAGGGUGCGUGGAAAACCCUUUAUGGGAUACUCGCUCCUCUUGCUGAUGACCAUGCGCACCU .((((.....)))).(((((........)))))((..((((((((...)))))))).))(((((((((((....(((.....)))......((.......))......)))))))).))) ( -35.70) >DroPer_CAF1 5538 120 - 1 GCGUGCAGAUCUCAAAGCUGAGUUUAACCAGCUGGCUGUUGUUUAUGGUCAGAGACUUGAGGGUCCGAGGGAAGCCCUUGAUGGGGUAUUCACUGCGCUUGCUGACCACCAUCCUCUGCU ....(((((((((..(((((........)))))((((((.....)))))).))))..((..((((..(((((.(((((....))))).))).))((....)).))))..))...))))). ( -38.80) >consensus GCGUGCAAAUCUCAAAGCUGAGCUUCACCAACUGGAUAUUGUUGAUGGUCAGCGAUUUGAGGGUGCGCGGAAAACCCUUGAUGGGAUACUCGCUGCGCUUGCUGAUUACCAUGCGCAGCU (((((((..(((((((.((((.(.(((.((((.....)))).))).).))))...)))))))....((((....(((.....)))....))))..................))))).)). (-21.27 = -21.97 + 0.70)

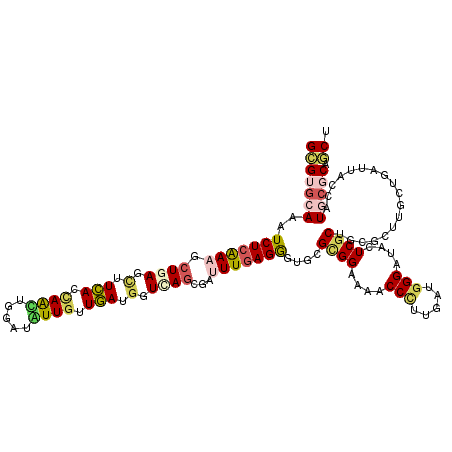
| Location | 10,517,911 – 10,518,020 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.51 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -29.22 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10517911 109 - 22407834 --GAUAACUAAUUAGUCAAUCCACCUUGGUCAGCUUGUUACCGCUGACAUCCAGCUUUGUGAGGUUGCGCAGGGUGCAUAUCUCGAAGCUCAGCUUCACUAGCUGGCUGUU --..........................((((((........))))))..((((((..(((((((((.((.(((.......)))...)).))))))))).))))))..... ( -36.80) >DroSec_CAF1 6245 109 - 1 --GAUAACUUAAUGGUCAAUCCACCUUGGUCAGCUUGUUACCGCUGACAUCCAGCUUGGUGAGGUUGCGCAAGGUGCAUAUCUCGAAGCUCAGCUUCACCAGCUGGCUGUU --(.((((....((..(((......)))..))....)))).)...((((.((((((.((((((((((.((.((((....))))....)).)))))))))))))))).)))) ( -40.30) >DroSim_CAF1 6252 109 - 1 --GAUAACAUAAUGGUCAAUCCACCUUGGUCAGCUUGUUACCGCUGACAUCCAGCUUGGUGAGGUUGCGCAGGGUGCAUAUCUCGAAGCUCAGCUUCACUAGCUGGCUGUU --(.(((((...((..(((......)))..))...))))).)...((((.((((((.((((((((((.((.(((.......)))...)).)))))))))))))))).)))) ( -41.70) >DroEre_CAF1 6371 109 - 1 --GAUAACUUAUUGGCCAACCCACCUUGGUCAGCUUGUUACCGCUGAUAUCCAGCUUUGUGAGGUUGCGCAGGGUGCAUAUCUCGAAGCUCAGCUUCACCAGCUGGCUGUU --(.((((...((((((((......))))))))...)))).)...((((.((((((..(((((((((.((.(((.......)))...)).))))))))).)))))).)))) ( -38.70) >DroYak_CAF1 6430 109 - 1 --GAUAACUUAUUGGUCAAUCCACCUUGGUCAGCUUGUUACCGCUGACAUCCAGCUUUGUGAGGUUGCGCAGGGUGCAUAUUUCGAAGCUCAGCUUCACCAGCUGGCUGUU --(.((((...(((..(((......)))..)))...)))).)...((((.((((((..(((((((((.((.(((.......)))...)).))))))))).)))))).)))) ( -35.50) >DroMoj_CAF1 8575 110 - 1 UUGA-AAUUGAUUUGUAUGGUAACCUGGGUUAGCUUGUUGCCGCUGAUAUCGAGCGUUGUUAGGUUGCGCAGCGUGCAAAUCUCGAAGCUCAGCUUCACCAAUUGAAUAUU ....-....(((((((((((((((((((....(((((.............)))))....)))))))))....))))))))))..(((((...))))).............. ( -30.92) >consensus __GAUAACUUAUUGGUCAAUCCACCUUGGUCAGCUUGUUACCGCUGACAUCCAGCUUUGUGAGGUUGCGCAGGGUGCAUAUCUCGAAGCUCAGCUUCACCAGCUGGCUGUU ............................((((((........))))))..(((((...(((((((((.((.(((.......)))...)).)))))))))..)))))..... (-29.22 = -29.58 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:46 2006