| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,494,388 – 10,494,491 |
| Length | 103 |
| Max. P | 0.992250 |

| Location | 10,494,388 – 10,494,491 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.23 |
| Mean single sequence MFE | -27.16 |
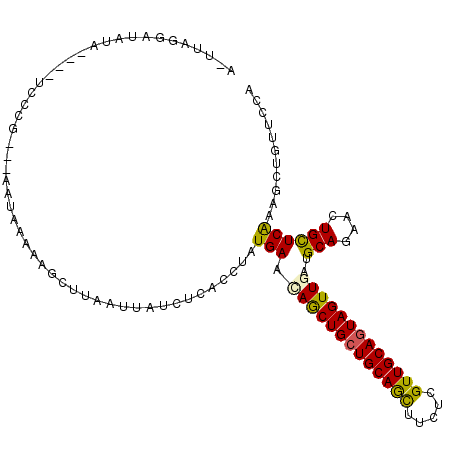
| Consensus MFE | -19.26 |
| Energy contribution | -18.68 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10494388 103 + 22407834 A-UUAUGAUAUA----UCGCU---AAUAAAAGAAUCAAGCAUAAUACCUAUGAAUAACUGCUGCAAUUUCUCGUUGCAAUAGUUGAUGCAGAACUGUUCAAAGCUGUUGCU (-(((((.....----...((---......)).......)))))).........((((((.((((((.....)))))).))))))..((((..((......))...)))). ( -16.66) >DroVir_CAF1 6805 103 + 1 --UUAGAG-AUACU-UUCCUGGGCCACU----GCUUAGUUAACUCACCAAUGAACAGCUGCUGCAGCUUCUCGUUGCAGUAGUUAAUGCAGAACUGCUCAAAGCUGUUCGA --...(((-....(-(..((((((....----))))))..))))).....(((((((((((((((((.....)))))))).......(((....)))....))))))))). ( -34.40) >DroEre_CAF1 5466 104 + 1 A-UUAUGAUAUU---AUCCCU---AAUAACCGUUUGCUUUAUCUUACCUAUGAAUAACUGCUGCAGUUUCUCGUUGCAGUAGUUGAUGCAGAACUGCUCAAAGCUAUUGCA .-..........---......---.......((..(((((...............(((((((((((.......)))))))))))((.(((....))))))))))....)). ( -22.80) >DroYak_CAF1 5415 103 + 1 A-UUACAAUAUA---AA-UCG---AAUAACAAACUUCACUAUCAUACCUAUGAACAGCUGCUGCAGUUUCUCGUUGCAGUAGUUGAUGCAGAACUGCUCGAAGCUGUUGCA .-..........---..-...---..(((((..((((....((((....)))).((((((((((((.......))))))))))))..(((....)))..)))).))))).. ( -26.40) >DroMoj_CAF1 6175 106 + 1 --UUAGGUUCUACUCCUCCAGUGGCAUUAGACGCU---UCCGCUCACCUAUGAACAGCUGCUGCAGCUUCUCGUUGCAGUAGUUGAUGCAGAACUGCUCAAAGCUAUUCGA --.(((((.(((((.....))))).....((.((.---...)))))))))....(((((((((((((.....))))))))))))).....(((..((.....))..))).. ( -33.50) >DroPer_CAF1 6094 104 + 1 AAUCAGCAUGAA----UCAAG---GAUAGAAAGCUUUCCACACUCACCAAUGAACAGCUGCUGCAGCUUCUCGUUGCAGUAGUUAAUGCAGAACUGCUCGAAACUGUUCGA ............----....(---((.((....)).)))...........(((((((((((((((((.....)))))))))).....(((....)))......))))))). ( -29.20) >consensus A_UUAGGAUAUA____UCCCG___AAUAAAAAGCUUAAUUAUCUCACCUAUGAACAGCUGCUGCAGCUUCUCGUUGCAGUAGUUGAUGCAGAACUGCUCAAAGCUGUUCCA ..................................................(((.(((((((((((((.....)))))))))))))..(((....))))))........... (-19.26 = -18.68 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:43 2006