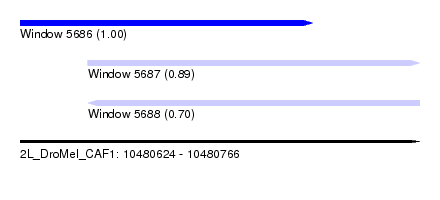
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,480,624 – 10,480,766 |
| Length | 142 |
| Max. P | 0.998997 |

| Location | 10,480,624 – 10,480,728 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10480624 104 + 22407834 ----------------CCUGUUUUAUUAUGGACUUUUGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACCAACAAAAUACUACUGGUUCGCAGCCAAUUUUGU ----------------..(((((((.(((((((....)))))))...(((....)))))))))).(((((((.(..((((((((.............))))))))..))))))))..... ( -33.32) >DroSec_CAF1 8767 118 + 1 -CAUGGCACCU-CCUACCUGUUUUAUUAUGGACUUUUGUCCAUGUUUGCUUCCUGGCUAGAACAUAUUGCCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGU -..((((....-.((((((((((((.(((((((....)))))))...(((....))))))))))........)))))(((((((.............))))))).....))))....... ( -33.42) >DroSim_CAF1 9095 119 + 1 -CAUGGCACCUUCUUACCUGUUUUAUUAUGGACUUUUGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGU -..(((((((..((....(((((((.(((((((....)))))))...(((....))))))))))....))..))).((((((((.............))))))))....))))....... ( -32.02) >DroEre_CAF1 12636 117 + 1 ACAUGUUACCU-CCUACCUGUUUUAUUA--GACUUUUGUCUAUGUUUGCUUUCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAGAAUUCUGCUGGUUCGAAGCCAAUUUUGU ..(((((....-((.....((.....((--(((....))))).....)).....))....)))))(((((((....((((((((.............))))))))...)))))))..... ( -29.42) >DroYak_CAF1 9477 119 + 1 ACAUAGCACCU-CCUACCUGUUUAAUUAUGGACUUUUGUCUAUGUUUGCUUCCUGGCUAAAACCUAUUGGCUGUUGGACCAGUAACUAACACAAUUCUGCUGGUUCAAAGCCAAUUUUGU ...........-.......((((...(((((((....)))))))...(((....)))..))))..(((((((.(((((((((((.............))))))))))))))))))..... ( -31.62) >consensus _CAUGGCACCU_CCUACCUGUUUUAUUAUGGACUUUUGUCUAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGU ..................(((((((.(((((((....)))))))...(((....)))))))))).(((((((....((((((((.............))))))))...)))))))..... (-27.00 = -27.56 + 0.56)

| Location | 10,480,648 – 10,480,766 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10480648 118 + 22407834 UAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACCAACAAAAUACUACUGGUUCGCAGCCAAUUUUGUAGUUUUU--GUUUUCCUUUUAGUGUGCAACAAGGAAAAAA ((((((((((....)))..)))))))((((((.(..((((((((.............))))))))..))))))).............--.((((((((...((.....)))))))))).. ( -32.82) >DroSec_CAF1 8805 120 + 1 CAUGUUUGCUUCCUGGCUAGAACAUAUUGCCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGUGUUUUCCUUUUCGUGUGCAACAAGGAAAAAC .(((((((((....)))..))))))(((((.(((....))))))))..((((((.(((((((((.....)))).....))))))))))).((((((((...((.....)))))))))).. ( -27.80) >DroSim_CAF1 9134 119 + 1 UAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAAUUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUUGUGUUUUCCUUUUCGUGUGCAACAAGGAA-AAC ((((((((((....)))..)))))))...(((((....))))).....((((((.(((((((((.....)))).....)))))))))))(((((((((...((.....))))))))-))) ( -32.00) >DroEre_CAF1 12673 117 + 1 UAUGUUUGCUUUCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAGAAUUCUGCUGGUUCGAAGCCAAUUUUGUAAUUUUU--GUUUUCCUUUUCAUGUGCAACAAGGAA-AAC ((((((((((....)))..)))))))((((((....((((((((.............))))))))...)))))).............--(((((((((............))))))-))) ( -30.82) >DroYak_CAF1 9516 117 + 1 UAUGUUUGCUUCCUGGCUAAAACCUAUUGGCUGUUGGACCAGUAACUAACACAAUUCUGCUGGUUCAAAGCCAAUUUUGUAGUUUUU--GUUUUCCUUUUCGUGUGCAACAAGGAU-AAC ..............(((((......(((((((.(((((((((((.............))))))))))))))))))....)))))...--(((.(((((...((.....))))))).-))) ( -30.42) >consensus UAUGUUUGCUUCCUGGCUAGAACAUAUUGGCUGGUAGACCAGUAACUAACAAAAUACUGCUGGUUCGAAGCCAAUUUUGUAGUUUUU__GUUUUCCUUUUCGUGUGCAACAAGGAA_AAC .(((((((((....)))..))))))(((((((....((((((((.............))))))))...))))))).................((((((...((.....)))))))).... (-23.74 = -24.22 + 0.48)

| Location | 10,480,648 – 10,480,766 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
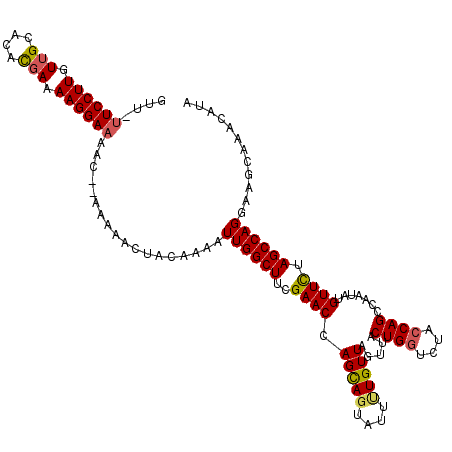
>2L_DroMel_CAF1 10480648 118 - 22407834 UUUUUUCCUUGUUGCACACUAAAAGGAAAAC--AAAAACUACAAAAUUGGCUGCGAACCAGUAGUAUUUUGUUGGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGGAAGCAAACAUA ..((((((((............)))))))).--............((((((((...((((((((...........))))))))....))))))))(((((...((......)).))))). ( -28.30) >DroSec_CAF1 8805 120 - 1 GUUUUUCCUUGUUGCACACGAAAAGGAAAACACAAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAAUUACUGGUCUACCAGGCAAUAUGUUCUAGCCAGGAAGCAAACAUG ((((((((((.(((....))).)))))))).)).............((((((..((((.(((((....))))).....((((....)))).......)))).))))))............ ( -27.60) >DroSim_CAF1 9134 119 - 1 GUU-UUCCUUGUUGCACACGAAAAGGAAAACACAAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAAUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGGAAGCAAACAUA (((-((((((.(((....))).)))))))))...............((((((..((((.(((((....))))).....((((....)))).......)))).))))))............ ( -29.60) >DroEre_CAF1 12673 117 - 1 GUU-UUCCUUGUUGCACAUGAAAAGGAAAAC--AAAAAUUACAAAAUUGGCUUCGAACCAGCAGAAUUCUGUUAGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGAAAGCAAACAUA (((-((((((.((......)).)))))))))--.............((((((..((((.(((((....))))).....((((....)))).......)))).))))))............ ( -30.50) >DroYak_CAF1 9516 117 - 1 GUU-AUCCUUGUUGCACACGAAAAGGAAAAC--AAAAACUACAAAAUUGGCUUUGAACCAGCAGAAUUGUGUUAGUUACUGGUCCAACAGCCAAUAGGUUUUAGCCAGGAAGCAAACAUA (((-.(((((.(((....))).))))).)))--............((((((((((.(((((...(((((...))))).))))).))).))))))).(((....))).............. ( -28.50) >consensus GUU_UUCCUUGUUGCACACGAAAAGGAAAAC__AAAAACUACAAAAUUGGCUUCGAACCAGCAGUAUUUUGUUAGUUACUGGUCUACCAGCCAAUAUGUUCUAGCCAGGAAGCAAACAUA ....((((((.(((....))).))))))..................((((((..((((.(((((....))))).....((((....)))).......)))).))))))............ (-20.26 = -20.46 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:38 2006