| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,126,937 – 1,127,056 |
| Length | 119 |
| Max. P | 0.655428 |

| Location | 1,126,937 – 1,127,056 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
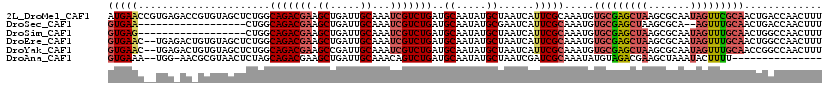
>2L_DroMel_CAF1 1126937 119 + 22407834 AUGAACCGUGAGACCGUGUAGCUCUGGCAGACGAAGCUGAUUGCAAAUCGUCUGAUGCAAUAUGCUAAUCAUUCGCAAAUGUGCGAGCUAAGCGCAAUAGUUCGCAACUGACCAACUUU ..(((((....)..(((.((((((...(((((((.((.....))...)))))))..(((...(((.........)))....))))))))).))).....))))................ ( -32.70) >DroSec_CAF1 13551 99 + 1 GUGAA------------------CUGGCAGACGAAGCUGAUUGCAAAUCGUCUGAUGCAAUAUGCGAAUCAUUCGCAAAUGUGCGAGCUAAGCGCA--AGUUUGCAACUGACCAACUUU .....------------------.((((((((((.((.....))...))))))).(((((..((((.....((((((....)))))).....))))--...))))).....)))..... ( -30.90) >DroSim_CAF1 13384 101 + 1 GUGAG------------------CUGGCAGACGAAGCUGAUUGCAAAUCGUCUGAUGCAAUAUGCUAAUCAUUCGCAAAUGUGCGAGCUAAGCGCAAUAGUUUGCAACUGGCCAACUUU ((..(------------------(..((((((((.((.....))...))))))).(((((...((((.....(((((....)))))((.....))..))))))))).)..))..))... ( -31.10) >DroEre_CAF1 13987 117 + 1 GUGAAC--UGAGACUGUGUAGCUCUGGCAGACGAAGCUGAUUGCAAAUCGUCUGAUGCAAUAUGCUAAUCAUUCGCAAAUGUGCGAGCUAAGCGCAAUAGUUUGCAACUGGCCAACUUU (..(((--((....((((((((((...(((((((.((.....))...)))))))..(((...(((.........)))....))))))))..))))).)))))..).............. ( -34.70) >DroYak_CAF1 14245 117 + 1 GUGAAC--UGAGACUGUGUAGCUCUGGCAGACGAAGCCGAUUGCAAAUCGUCUGAUGCAAUAUGCUAAUCAUUCGCAAAUGUGCGAGCUAAGCGCAAUAGUUUGCAACCGGCCAACUUU (..(((--((....((((((((((...(((((((.((.....))...)))))))..(((...(((.........)))....))))))))..))))).)))))..).............. ( -33.50) >DroAna_CAF1 14157 101 + 1 GUGAAA--UGG-AACGCGUAACUCUAGCAGACGAAGCUGAUUGCAAACAGUCUGAUGCAAUAUGCUAAUCGAUCGCAAAUAUGUAGACGAAGCUAAAUACUUUU--------------- (((...--...-..)))(((....((((......(((..((((((..........))))))..)))..(((.(((((....))).))))).))))..)))....--------------- ( -16.40) >consensus GUGAAC__UGA_AC_GUGUAGCUCUGGCAGACGAAGCUGAUUGCAAAUCGUCUGAUGCAAUAUGCUAAUCAUUCGCAAAUGUGCGAGCUAAGCGCAAUAGUUUGCAACUGGCCAACUUU (((((......................(((((((.((.....))...)))))))..((.....))......))))).....(((((((((.......)))))))))............. (-20.85 = -21.85 + 1.00)
| Location | 1,126,937 – 1,127,056 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -18.67 |
| Energy contribution | -20.42 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
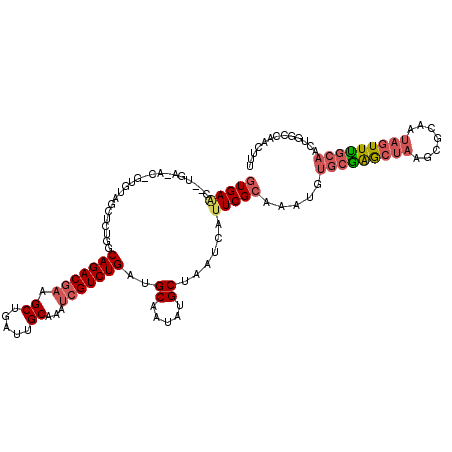
>2L_DroMel_CAF1 1126937 119 - 22407834 AAAGUUGGUCAGUUGCGAACUAUUGCGCUUAGCUCGCACAUUUGCGAAUGAUUAGCAUAUUGCAUCAGACGAUUUGCAAUCAGCUUCGUCUGCCAGAGCUACACGGUCUCACGGUUCAU ...((((((((.(((((((....((((.......))))..))))))).)))))))).........(((((((...((.....)).)))))))...((((....((......)))))).. ( -33.10) >DroSec_CAF1 13551 99 - 1 AAAGUUGGUCAGUUGCAAACU--UGCGCUUAGCUCGCACAUUUGCGAAUGAUUCGCAUAUUGCAUCAGACGAUUUGCAAUCAGCUUCGUCUGCCAG------------------UUCAC ...((..((((.(((((((..--((((.......))))..))))))).))))..))..((((...(((((((...((.....)).))))))).)))------------------).... ( -30.00) >DroSim_CAF1 13384 101 - 1 AAAGUUGGCCAGUUGCAAACUAUUGCGCUUAGCUCGCACAUUUGCGAAUGAUUAGCAUAUUGCAUCAGACGAUUUGCAAUCAGCUUCGUCUGCCAG------------------CUCAC ..(((((((..(.(((((.....(((..(((..(((((....))))).)))...)))..))))).)((((((...((.....)).)))))))))))------------------))... ( -33.10) >DroEre_CAF1 13987 117 - 1 AAAGUUGGCCAGUUGCAAACUAUUGCGCUUAGCUCGCACAUUUGCGAAUGAUUAGCAUAUUGCAUCAGACGAUUUGCAAUCAGCUUCGUCUGCCAGAGCUACACAGUCUCA--GUUCAC .....((((..(.(((((.....(((..(((..(((((....))))).)))...)))..))))).)((((((...((.....)).))))))))))(((((..........)--)))).. ( -31.80) >DroYak_CAF1 14245 117 - 1 AAAGUUGGCCGGUUGCAAACUAUUGCGCUUAGCUCGCACAUUUGCGAAUGAUUAGCAUAUUGCAUCAGACGAUUUGCAAUCGGCUUCGUCUGCCAGAGCUACACAGUCUCA--GUUCAC ..((..(((((((((((((.....((.....))(((((....))))).((((..((.....)))))).....)))))))))))))....))....(((((..........)--)))).. ( -32.30) >DroAna_CAF1 14157 101 - 1 ---------------AAAAGUAUUUAGCUUCGUCUACAUAUUUGCGAUCGAUUAGCAUAUUGCAUCAGACUGUUUGCAAUCAGCUUCGUCUGCUAGAGUUACGCGUU-CCA--UUUCAC ---------------....(((((((((.((((..........))))..(((.(((..((((((..........))))))..)))..))).))))))..))).....-...--...... ( -17.50) >consensus AAAGUUGGCCAGUUGCAAACUAUUGCGCUUAGCUCGCACAUUUGCGAAUGAUUAGCAUAUUGCAUCAGACGAUUUGCAAUCAGCUUCGUCUGCCAGAGCUACAC_GU_UCA__GUUCAC ....(((((..(.(((((.....(((.......(((((....))))).......)))..))))).)((((((...((.....)).)))))))))))....................... (-18.67 = -20.42 + 1.75)

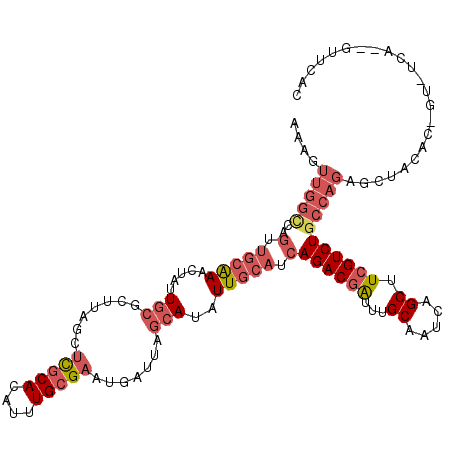
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:54 2006