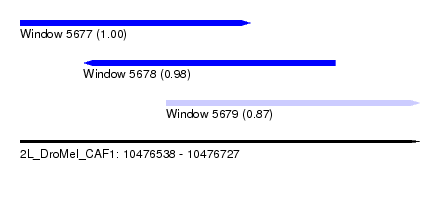
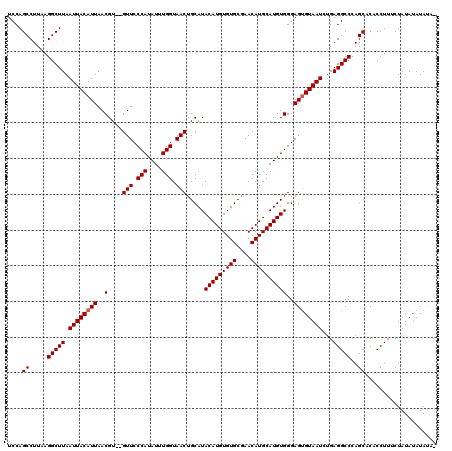
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,476,538 – 10,476,727 |
| Length | 189 |
| Max. P | 0.995727 |

| Location | 10,476,538 – 10,476,647 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.30 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10476538 109 + 22407834 UUACCAACCUGUAUAUAACAUUU--------AUAUGUA---UAUAUAUAUAGAAAGGUGUGCUGGGCCUCCGAUUACACUCCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAU ........(((((((((((((..--------..)))).---..)))))))))...((((.((((((...)).............(((((((((......))))))))))))))))).... ( -26.20) >DroSec_CAF1 4922 112 + 1 UUACCAACCUGUAUAUAACAUAU--------AUAUGUAGGGUAUAUAUAUAGAAAGGUGUGCUGGGCCUCAGAUUACACUCCCACAUGCAUGUUAGCACACAUGUAUGCAGUUACCAAAU ........(((((((((...(((--------(((((((...)))))))))).....((((((((((....((......)))))(((....))).))))))).)))))))))......... ( -27.20) >DroSim_CAF1 4683 112 + 1 UUACCAACCUGUAUAUAACAUAC--------AUAUGUAGGGUAUAUAUAUAGAAAGGUGUGCUGGGCCUCAGAUUACACUCCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAU ........(((((((((((.(((--------....)))..)).)))))))))...((((.((((((....((......))))).(((((((((......))))))))).))))))).... ( -27.20) >DroEre_CAF1 5289 116 + 1 UUACCGAGCUGUAUAUAAAGUAUACAUAUAUAUAU--GA--AGUAUAUAUAGAAAGGUGUGCUGGGCCUCAUAUUACACUCCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAU ......(((((((((((.........(((((((((--..--.))))))))).....((((((.(((..............)))(((....)))..)))))).)))))))))))....... ( -27.94) >DroYak_CAF1 5327 108 + 1 UUACCAACCUGUAUAUAAAGUAU--------AUAU--AG--AGUAUAUAUAGAAAGGUGCGCUGGGCCUCAUAUUACUCUCCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAU ......(((((((((((...)))--------))))--))--.))...........((((.((((((..............))).(((((((((......))))))))).))))))).... ( -23.94) >consensus UUACCAACCUGUAUAUAACAUAU________AUAUGUAG__UAUAUAUAUAGAAAGGUGUGCUGGGCCUCAGAUUACACUCCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAU ........((((((((((((((((......)))))))......)))))))))...((((.((((((..............))).(((((((((......))))))))).))))))).... (-23.14 = -24.30 + 1.16)

| Location | 10,476,568 – 10,476,687 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.95 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10476568 119 - 22407834 UCCAGCCUUAAGGCUUAAUUACAUUAACGUGUGUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCGAACAUGCAUGUGGGAGUGUAAUCGGAGGCCCAGCACACCUUUCUAUAUAUAUA- (((((((....))))..((((((((..((((((((.(((....))).))).))))(((((((((....))))))))))..)))))))).)))...........................- ( -31.80) >DroSec_CAF1 4954 118 - 1 UCCAGCCUUAAGGCUUAAUUACAUUAACGU--GUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCUAACAUGCAUGUGGGAGUGUAAUCUGAGGCCCAGCACACCUUUCUAUAUAUAUAC ....((.....(((((.((((((((..(((--(((.(((....))).))).))..(((((((((....))))))))))..))))))))...)))))..)).................... ( -29.70) >DroSim_CAF1 4715 118 - 1 UCCAGCCUUAAGGCUUAAUUACAUUAACGU--GUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCGAACAUGCAUGUGGGAGUGUAAUCUGAGGCCCAGCACACCUUUCUAUAUAUAUAC (((((((....))))....(((((...(((--(((((((....))).....(((((....)))))))))))).))))))))((((...(((.....))).))))................ ( -29.80) >DroEre_CAF1 5326 117 - 1 UCCAGCCUUAAGGCUUAAUUACAUUAACGU--GUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCGAACAUGCAUGUGGGAGUGUAAUAUGAGGCCCAGCACACCUUUCUAUAUAUACU- (((((((....))))....(((((...(((--(((((((....))).....(((((....)))))))))))).))))))))((((((((.((((.........))))..))).))))).- ( -30.60) >DroYak_CAF1 5356 117 - 1 UCCAGCCUUAAGGCUUAAUUACAUUAACGU--GUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCGAACAUGCAUGUGGGAGAGUAAUAUGAGGCCCAGCGCACCUUUCUAUAUAUACU- ....((((((..((((..((((((...(((--(((((((....))).....(((((....)))))))))))).))))))..))))....))))))........................- ( -30.90) >consensus UCCAGCCUUAAGGCUUAAUUACAUUAACGU__GUUCCCAUAUUUGGUAACUGCAUACAUGUGUGCGAACAUGCAUGUGGGAGUGUAAUCUGAGGCCCAGCACACCUUUCUAUAUAUAUA_ ....((.....(((((.((((((((..(....(((.(((....))).))).....(((((((((....))))))))))..))))))))...)))))..)).................... (-27.90 = -28.10 + 0.20)

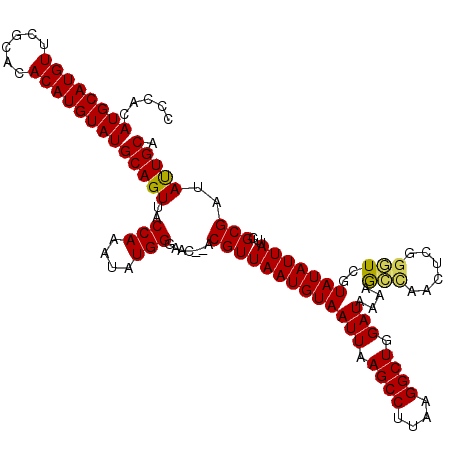
| Location | 10,476,607 – 10,476,727 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10476607 120 + 22407834 CCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAUAUGGGAACACACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAUGCCAACUCGAGUCGUAUAUUAUUUGCGAUAUUGCA ......((((...(((((.....((((((.(((.(((....))).)))((.((.....((((((.((((....)))).)))...))).....)).)).))))))....)))))...)))) ( -28.00) >DroSec_CAF1 4994 118 + 1 CCCACAUGCAUGUUAGCACACAUGUAUGCAGUUACCAAAUAUGGGAAC--ACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAAGCCAACUCGGGUCGUAUAUUAUUCGCGAUAUUGCA .....((((((((......))))))))((((((.(((....))).)))--.((((((((((.....((((...((((.......)))).....))))..)))))))...)))....))). ( -27.90) >DroSim_CAF1 4755 118 + 1 CCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAUAUGGGAAC--ACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAAGCCAACGCGGGUCGUAUAUUAUUCGCGAUAUUGCA ....(((((((((......))))))))).((((((..((..((....)--)..))...))))))..((..((.((((.......))))..(((((((........))))))).))..)). ( -30.30) >DroEre_CAF1 5365 118 + 1 CCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAUAUGGGAAC--ACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAAAGUAACUCGGGUCAUAUAUUAUACGCGAUAUUGCA (((.(((((((((......))))))))).((((((..((..((....)--)..))......(((.((((....)))).))).....)))))).))).............(((....))). ( -28.70) >DroYak_CAF1 5395 118 + 1 CCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAUAUGGGAAC--ACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAAAGUAACUCGGCUCGUAUAUUAUACGCGAUACUGCA .....((((((((......))))))))(((((..(((....)))...(--.((((((((((....((((.....(((........))).....))))..))))))).))).)..))))). ( -30.40) >consensus CCCACAUGCAUGUUCGCACACAUGUAUGCAGUUACCAAAUAUGGGAAC__ACGUUAAUGUAAUUAAGCCUUAAGGCUGGAUAAAAGCCAACUCGGGUCGUAUAUUAUUCGCGAUAUUGCA .....((((((((......))))))))(((((..(((....))).......(((((((((((((.((((....)))).)))....(((......)))..)))))))...)))..))))). (-24.80 = -24.40 + -0.40)

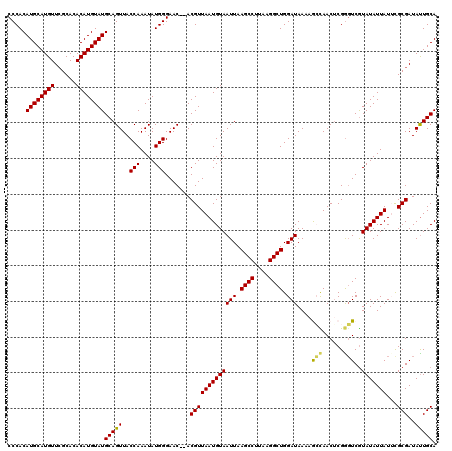
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:28 2006