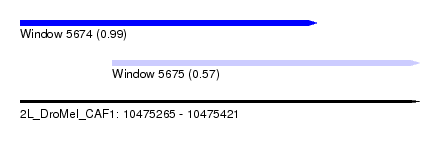
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,475,265 – 10,475,421 |
| Length | 156 |
| Max. P | 0.989061 |

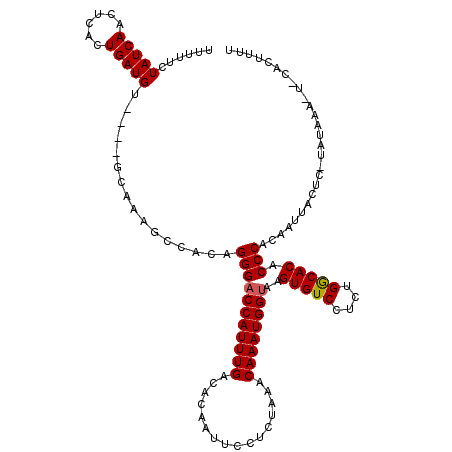
| Location | 10,475,265 – 10,475,381 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -19.37 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10475265 116 + 22407834 UUUUUCUAUCAACUCACUGAUGU----GCAAAGCCACAGGGACCAUUUGACACAAUUCCCCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAAUUACUCGUAUAAAAUCCACUUUU .......((((......))))((----(......))).(((((((((((................))))))))..(((((....))))).)))........................... ( -20.59) >DroSec_CAF1 3748 103 + 1 UUUUUCUAUCAACUCACUGAUGU----GCAAAGCCACUGGGACCAUUUGACACAAUUCCUCUAAACAAAUGGUAAGUGUCCUCCGGCACACCCACAAUUA-------------CACUUUU ......(((((......))))).----..........((((((((((((................))))))))..(((((....))))).))))......-------------....... ( -20.49) >DroSim_CAF1 3498 103 + 1 UUUUUCUAUCAACUCACUGAUGU----GCAAAGCCACAGGGCCCAUUUGACACAAUUCCUCUAAACAAAUGGUAAGUGUCCUCCGGCACACCCACAAUUA-------------CACUUUU .......((((......))))((----(....(((..((((((((((((................))))))).....)))))..))).....))).....-------------....... ( -18.39) >DroEre_CAF1 3951 120 + 1 UUUUUCUAUCAACUCACUGAUGUACAUGCAAAGCCACAGGGACCAUUUGACACAAUUUCUCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAUUUACUCAUAUAAAUUGAACUUUG ......(((((......)))))......(((((.....(((((((((((................))))))))..(((((....))))).)))........(((.......))).))))) ( -22.69) >DroYak_CAF1 3984 120 + 1 UUUUUCUAUCAACACACUGAUGUGCUUGCAAAGCCACAGGGACCAUUUGACACAAUUCCUCUUAACAAAUGGUAAGUGUCCUCUGACACACCCACACUUACUCAUAUAAAAUAAACAUUG ..................((((((((.....)))....(((((((((((................))))))))..(((((....))))).))).....................))))). ( -22.79) >consensus UUUUUCUAUCAACUCACUGAUGU____GCAAAGCCACAGGGACCAUUUGACACAAUUCCUCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAAUUACUC_UAUAAA_U_CACUUUU ......(((((......)))))................(((((((((((................))))))))..(((((....))))).)))........................... (-19.37 = -19.41 + 0.04)

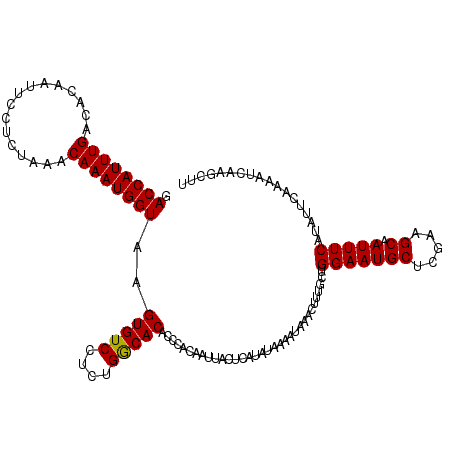
| Location | 10,475,301 – 10,475,421 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.32 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10475301 120 + 22407834 GACCAUUUGACACAAUUCCCCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAAUUACUCGUAUAAAAUCCACUUUUUCGGAAUGCUCGAAGCAAUUUCAUAUUCAAAAUCAUACUU .((((((((................))))))))..(((((....)))))..............((((....(((........))).(((.....))).................)))).. ( -19.09) >DroEre_CAF1 3991 120 + 1 GACCAUUUGACACAAUUUCUCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAUUUACUCAUAUAAAUUGAACUUUGCUGGAAUGCUCGAAGCAAUUUCAUACUCAACAUCAAGCUC .((((((((................))))))))..(((((....)))))......................((((..(((((.((....))..))))).))))................. ( -19.79) >DroYak_CAF1 4024 120 + 1 GACCAUUUGACACAAUUCCUCUUAACAAAUGGUAAGUGUCCUCUGACACACCCACACUUACUCAUAUAAAAUAAACAUUGCUGGAAUGCUCGAGGCAAUUUCAUAUUCAAAAUCAAGCUU .((((((((................))))))))..(((((....)))))...........................((((((.((....))..))))))..................... ( -18.49) >consensus GACCAUUUGACACAAUUCCUCUAAACAAAUGGUAAGUGUCCUCUGGCACACCCACAAUUACUCAUAUAAAAUAAACUUUGCUGGAAUGCUCGAAGCAAUUUCAUAUUCAAAAUCAAGCUU .((((((((................))))))))..(((((....))))).................................(((((((.....)).))))).................. (-17.54 = -17.32 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:24 2006