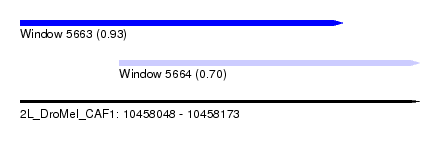
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,458,048 – 10,458,173 |
| Length | 125 |
| Max. P | 0.934726 |

| Location | 10,458,048 – 10,458,149 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.02 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -7.66 |
| Energy contribution | -9.18 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
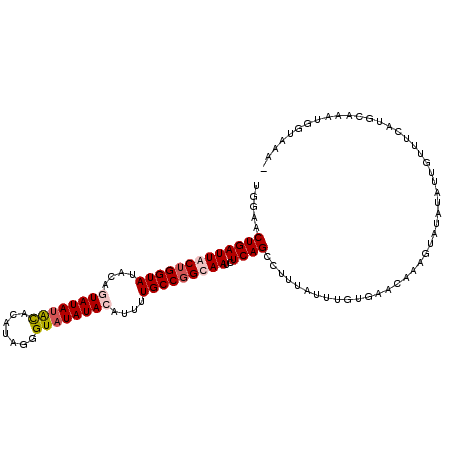
>2L_DroMel_CAF1 10458048 101 + 22407834 CACCUUCUACAGGUUCAGUUUAAAGUAAGAAUGGAACUGAUUACUGGUAUACAGUAUAAACGCAUAGGGUAUAUAAAU-UUGCCGGCAAUCUCAGCAUUAAU ...........(((((.((((.......)))).)))))((((.((((((....(((((..(.....)..)))))....-.)))))).))))........... ( -19.20) >DroSec_CAF1 7228 97 + 1 CACCUCAUACAGG-----UUUAAAGUAAGAAUGGAACUGAUUACUGGUAUACAGUAUAUGCACAUAGGGUAUAUACAUUUUGCCGGUAAUCUCAGCCUUUUU .((((.....)))-----).......(((((.((....(((((((((((....((((((((.......))))))))....)))))))))))....))))))) ( -28.40) >DroSim_CAF1 7012 97 + 1 CACCUUCUACAUG-----UUUAAAGUAAGAAUGGAACUGAUUACUGGUAUACAAUAUAUACACAUGGGGUAUAUACAUUUUGCCGGUAAUCUCAGCUUUUAU ..((((((((...-----......)).)))).))....(((((((((((.....(((((((.......))))))).....)))))))))))........... ( -23.40) >DroEre_CAF1 7896 82 + 1 CUC---CCACAGGUAGAGUU-AAAGUAAG-----AACUGCUUACUGC-----------UGUACCUAGGGUAUAUACAUUUUGCCGGCCAUCUCAGCCUUUAU ..(---((..(((((.(((.-..((((((-----.....))))))))-----------).))))).)))...............(((.......)))..... ( -21.60) >DroYak_CAF1 8245 93 + 1 -------UACAGGUCGAGUUUAAUGUGAGAAUGGAACUGAUUACGGGUACAAUGUAUAUAUACCCGGGGUAUAUACAAUUUGCCGGCAAACUCAGACUUU-- -------...(((((((((((..(((((............)))))((((.(.(((((((((.(....)))))))))).).))))...)))))).))))).-- ( -23.00) >consensus CACCU_CUACAGGU__AGUUUAAAGUAAGAAUGGAACUGAUUACUGGUAUACAGUAUAUACACAUAGGGUAUAUACAUUUUGCCGGCAAUCUCAGCCUUUAU ....................................((((...((((((.....(((((((.......))))))).....)))))).....))))....... ( -7.66 = -9.18 + 1.52)



| Location | 10,458,079 – 10,458,173 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -15.85 |
| Energy contribution | -16.98 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10458079 94 + 22407834 UGGAACUGAUUACUGGUAUACAGUAUAAACGCAUAGGGUAUAUAAAU-UUGCCGGCAAUCUCAGCAUUAAUUUGUGAACAA---------------AUGCAAAAGGUGAA- .......((((.((((((....(((((..(.....)..)))))....-.)))))).))))(((.(....(((((....)))---------------))......).))).- ( -19.60) >DroSec_CAF1 7254 110 + 1 UGGAACUGAUUACUGGUAUACAGUAUAUGCACAUAGGGUAUAUACAUUUUGCCGGUAAUCUCAGCCUUUUUUUGUGAGCGUAGUAUAUAUUGUUUCAUGCAAAUGGUAAA- .......(((((((((((....((((((((.......))))))))....)))))))))))...(((....((((((((((..........)))))...))))).)))...- ( -29.40) >DroSim_CAF1 7038 110 + 1 UGGAACUGAUUACUGGUAUACAAUAUAUACACAUGGGGUAUAUACAUUUUGCCGGUAAUCUCAGCUUUUAUUUGUGAAUAGAGUAUAUAUUGUUUCAUGCAAAUGGUAAA- ((((((.(((((((((((.....(((((((.......))))))).....)))))))))))...(((((.(((....)))))))).......)))))).............- ( -26.10) >DroYak_CAF1 8269 109 + 1 UGGAACUGAUUACGGGUACAAUGUAUAUAUACCCGGGGUAUAUACAAUUUGCCGGCAAACUCAGACUUU--UUCCGAACACAGUAUACUUCGUUUCAUGCAAAUAAUAAAA ((((((.......(((((...........)))))(((((((((....((((....)))).((.((....--.)).)).....))))))))))))))).............. ( -20.30) >consensus UGGAACUGAUUACUGGUAUACAGUAUAUACACAUAGGGUAUAUACAUUUUGCCGGCAAUCUCAGCCUUUAUUUGUGAACAAAGUAUAUAUUGUUUCAUGCAAAUGGUAAA_ .....(((((((((((((....((((((((.......))))))))....)))))))))..))))............................................... (-15.85 = -16.98 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:13 2006