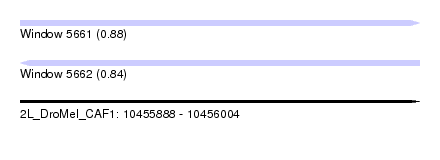
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,455,888 – 10,456,004 |
| Length | 116 |
| Max. P | 0.875308 |

| Location | 10,455,888 – 10,456,004 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10455888 116 + 22407834 UAUGU----ACUCCAGAAUGGGUUACCUCGAAUCCACAGGUUGUCUCGCAUAUCGCUGUGUUCAGUAGAAACAGUGUGAUAAAUACGAGUAUGCCACCGAAGUAUUCCAUUUCGGUAAAC .....----.........((((((......))))))..(((...((((..((((((..(((((....).))))..))))))....))))...)))((((((((.....)))))))).... ( -29.60) >DroSec_CAF1 5093 120 + 1 UAUGUAUCUACUUCAGAAUGGGUUACCUCGAAUCCACAGGUUGCCUCGCAUAUCGCUGUGUUCAGUAGAAACAGUGCGAUAAAUGCGAGUGUGCCACCGAAGUAUUCCAUUUCGGUGAAC ...........(((....((((((......))))))..(((...((((((((((((..(((((....).))))..))))))..))))))...)))((((((((.....))))))))))). ( -36.30) >DroSim_CAF1 4881 120 + 1 UAUGUAUAUACUUCCGAAUGGGUUACCUCGAAUCCACAGGUUGUCUCGCAUAUCGCUGUGUUCAGUAGAAACAGUGCGAUAAAUGCGAGUGUGCCACCCAAGUAUUCCAUUUCGGUGAAC .............((((((((..(((...(....)...(((...((((((((((((..(((((....).))))..))))))..))))))...)))......)))..))).)))))..... ( -33.90) >DroEre_CAF1 5767 116 + 1 ---UUAUGUACUUCA-AAUGCGUUACCUCAAAUCCACAGGUUGUCUCGCAUAUCGCUGUGUUCAGUAGAAACAGAGUGAUAAAAGCGAGGGUACCACUGAAGUAUUCCAUUUCGGUGGAC ---....((((((((-..((.(((......))).))..(((..((((((.(((((((.(((((....).)))).)))))))...))))))..)))..))))))))(((((....))))). ( -35.60) >DroYak_CAF1 6076 117 + 1 ---UUAGGUACUUCAGAAUGGGUUACCGCAAAUCCACAGGUCGUCUCGCAUAUCGCUGUGUUCAGUAGAAACAGAGUGAUAAAAGCGAGGGAACCACUGAAGUAUUCCAUUUCGGUGAAU ---...((((((((((..((((((......))))))..(((..((((((.(((((((.(((((....).)))).)))))))...))))))..))).))))))))..))............ ( -41.70) >DroAna_CAF1 7443 107 + 1 ---UUA----------AUCAAGUUACCUCGAAUCCACAGGUAGCCUCGGAUACCGAAGCAUACGGUAGCACUAAUGCGAUAAAUAUUAGCACUCUACCGAAAUAUUCCAUUUCAGUGAAC ---...----------.......(((((.(......))))))((.((((...)))).)).((((((((..((((((.......))))))....)))))(((((.....))))).)))... ( -21.60) >consensus ___GUAU_UACUUCAGAAUGGGUUACCUCGAAUCCACAGGUUGUCUCGCAUAUCGCUGUGUUCAGUAGAAACAGUGCGAUAAAUGCGAGUGUGCCACCGAAGUAUUCCAUUUCGGUGAAC ..................((((((......))))))..(((...(((((.((((((..(((((....))).))..))))))...)))))...)))((((((((.....)))))))).... (-22.42 = -23.48 + 1.06)

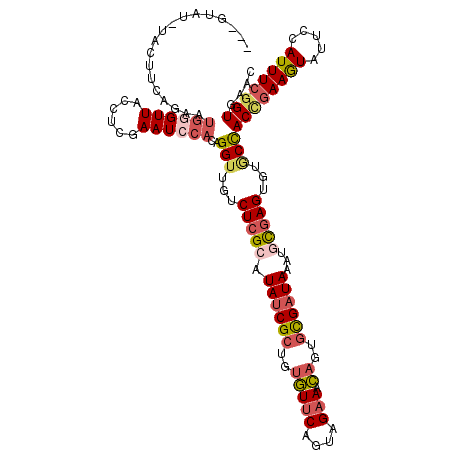
| Location | 10,455,888 – 10,456,004 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10455888 116 - 22407834 GUUUACCGAAAUGGAAUACUUCGGUGGCAUACUCGUAUUUAUCACACUGUUUCUACUGAACACAGCGAUAUGCGAGACAACCUGUGGAUUCGAGGUAACCCAUUCUGGAGU----ACAUA ((...(((.(((((..((((((((...((((((((((..((((...(((((((....))).)))).))))))))))......))))...))))))))..))))).)))...----))... ( -32.50) >DroSec_CAF1 5093 120 - 1 GUUCACCGAAAUGGAAUACUUCGGUGGCACACUCGCAUUUAUCGCACUGUUUCUACUGAACACAGCGAUAUGCGAGGCAACCUGUGGAUUCGAGGUAACCCAUUCUGAAGUAGAUACAUA .((((..(((.(((..((((((((...((((((((((..((((((..(((((.....)))))..))))))))))))......))))...))))))))..))))))))))........... ( -36.60) >DroSim_CAF1 4881 120 - 1 GUUCACCGAAAUGGAAUACUUGGGUGGCACACUCGCAUUUAUCGCACUGUUUCUACUGAACACAGCGAUAUGCGAGACAACCUGUGGAUUCGAGGUAACCCAUUCGGAAGUAUAUACAUA .....(((((.(((..(((((.((...((((((((((..((((((..(((((.....)))))..))))))))))))......))))...)).)))))..))))))))............. ( -37.50) >DroEre_CAF1 5767 116 - 1 GUCCACCGAAAUGGAAUACUUCAGUGGUACCCUCGCUUUUAUCACUCUGUUUCUACUGAACACAGCGAUAUGCGAGACAACCUGUGGAUUUGAGGUAACGCAUU-UGAAGUACAUAA--- .((((......)))).((((((((((.(((((((((...((((.((.(((((.....))))).)).)))).)))))....((...))......)))).)))...-))))))).....--- ( -32.00) >DroYak_CAF1 6076 117 - 1 AUUCACCGAAAUGGAAUACUUCAGUGGUUCCCUCGCUUUUAUCACUCUGUUUCUACUGAACACAGCGAUAUGCGAGACGACCUGUGGAUUUGCGGUAACCCAUUCUGAAGUACCUAA--- ............((..((((((((.((((..(((((...((((.((.(((((.....))))).)).)))).)))))..)))).((((.((......)).)))).))))))))))...--- ( -33.20) >DroAna_CAF1 7443 107 - 1 GUUCACUGAAAUGGAAUAUUUCGGUAGAGUGCUAAUAUUUAUCGCAUUAGUGCUACCGUAUGCUUCGGUAUCCGAGGCUACCUGUGGAUUCGAGGUAACUUGAU----------UAA--- ((((((.((((((...))))))(((((...((((((.........)))))).)))))(((.(((((((...))))))))))..))))))(((((....))))).----------...--- ( -32.20) >consensus GUUCACCGAAAUGGAAUACUUCGGUGGCACACUCGCAUUUAUCACACUGUUUCUACUGAACACAGCGAUAUGCGAGACAACCUGUGGAUUCGAGGUAACCCAUUCUGAAGUA_AUAA___ .........(((((..((((((((.......(((((...((((...(((((((....))).)))).)))).)))))((.....))....))))))))..)))))................ (-19.86 = -19.98 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:11 2006