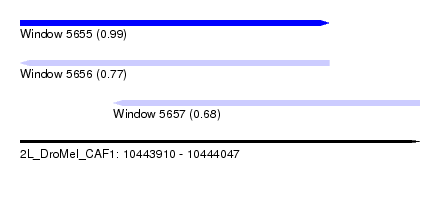
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,443,910 – 10,444,047 |
| Length | 137 |
| Max. P | 0.989750 |

| Location | 10,443,910 – 10,444,016 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
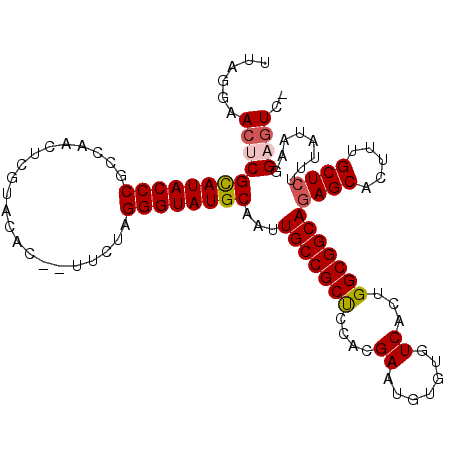
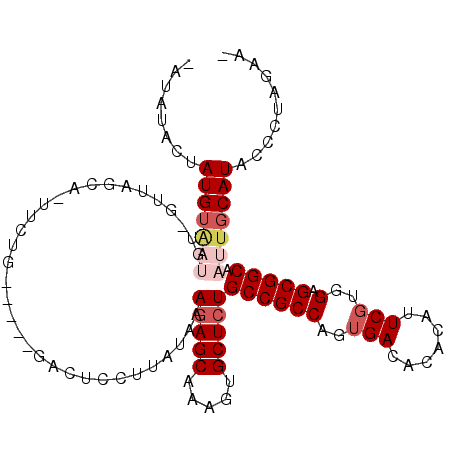
>2L_DroMel_CAF1 10443910 106 + 22407834 UUAGGAACUCGCAUACCCGCCAACUUGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAAUC- ..........((((((((..............--.....))))))))...(((((((.((((....)))).....)))))))((((.....)))).............- ( -32.11) >DroSec_CAF1 4294 106 + 1 UUAGGAACACGCAUACCCGCCAACUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAUAGCACUUUGCUCUUUAUAAGGAGUC- ..........((((((((..............--.....))))))))...(((((((.((((....)))).....))))))).........((((((....)))))).- ( -30.61) >DroSim_CAF1 4266 106 + 1 UUAGGAACACGCAUACCCGCCAACUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUC- ..........((((((((..............--.....))))))))...(((((((.((((....)))).....)))))))((((.....)))).............- ( -32.11) >DroEre_CAF1 4380 106 + 1 UUAGGAACUCGCAUACCCGCU-ACUCGUACAC--UUCUAGGGUAUGCAAUUGCCGCCCAAAGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUCAUAAGGAGUCU ...(((.(((((((((((...-..........--.....))))))))...(((((((....((.......))...)))))))((((.....))))........)))))) ( -35.67) >DroYak_CAF1 4353 108 + 1 UUAGGAACUCGUAUACCCGCU-ACUCACACACAGUUCUAGGGUAUGCAAUUGCCGCCCAACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUCU ...(((.(((.((((.(((((-(.(.(((((((.(((..((((..((....)).))))...)))))))))).).)))))).(((((.....))))).))))..)))))) ( -37.40) >consensus UUAGGAACUCGCAUACCCGCCAACUCGUACAC__UUCUAGGGUAUGCAAUUGCCGCUCCACGAAUGUGUGUCACUGGCGGCAGAGCACUUUGCUCUUUAUAAGGAGUC_ ......((((((((((((.....................))))))))...(((((((....((.......))...)))))))((((.....))))........)))).. (-29.40 = -29.80 + 0.40)

| Location | 10,443,910 – 10,444,016 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10443910 106 - 22407834 -GAUUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA--GUGUACAAGUUGGCGGGUAUGCGAGUUCCUAA -............(((((.....)))))((((((..(((.......)))..).)))))..((((((((((..((.--.(....)..))...))))))))))........ ( -32.80) >DroSec_CAF1 4294 106 - 1 -GACUCCUUAUAAAGAGCAAAGUGCUAUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA--GUGUACGAGUUGGCGGGUAUGCGUGUUCCUAA -.............(((((........(((((((..(((.......)))..).))))))...((((((((..((.--.(....)..))...)))))))).))))).... ( -30.00) >DroSim_CAF1 4266 106 - 1 -GACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA--GUGUACGAGUUGGCGGGUAUGCGUGUUCCUAA -.............(((((..((((.((((((((.((..(((......)))..)))))..(((.((((.......--)))).)))...)))))))))...))))).... ( -31.80) >DroEre_CAF1 4380 106 - 1 AGACUCCUUAUGAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCUUUGGGCGGCAAUUGCAUACCCUAGAA--GUGUACGAGU-AGCGGGUAUGCGAGUUCCUAA .(((((.......(((((.....)))))((((((...((.......))....))))))....((((((((.....--..........-...)))))))))))))..... ( -36.67) >DroYak_CAF1 4353 108 - 1 AGACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUUGGGCGGCAAUUGCAUACCCUAGAACUGUGUGUGAGU-AGCGGGUAUACGAGUUCCUAA .(((((..((((.(((((.....))))).((((.(.(.((((((((((...(((..((....))...)))..))).))))))).).)-.)))).)))).)))))..... ( -39.50) >consensus _GACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA__GUGUACGAGUUGGCGGGUAUGCGAGUUCCUAA .(((((.......(((((.....)))))((((((..(((.......)))..).)))))....((((((((.....................)))))))))))))..... (-28.12 = -28.96 + 0.84)


| Location | 10,443,942 – 10,444,047 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.64 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
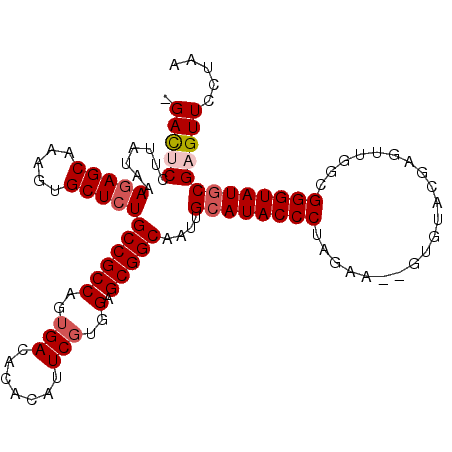
>2L_DroMel_CAF1 10443942 105 - 22407834 UAUGUACUAUGUUAUGUUGUUAGCAUUUCUG-----GAUUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA- ........((((((......))))))(((((-----(...........(((((.....)))))((((((..(((.......)))..).)))))...........))))))- ( -26.20) >DroSec_CAF1 4326 94 - 1 -AUAUACUAUGUAAUGU----------UCUG-----GACUCCUUAUAAAGAGCAAAGUGCUAUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA- -...............(----------((((-----(.(((........)))....((((..(((((((..(((.......)))..).))))))...))))...))))))- ( -22.70) >DroSim_CAF1 4298 94 - 1 -AUAUACUAUGUAAUGU----------UCUG-----GACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA- -......((((((((..----------....-----............(((((.....)))))((((((..(((.......)))..).))))).))))))))........- ( -24.10) >DroEre_CAF1 4411 101 - 1 -AUAU---AUGGG-----GUUAGCAGUUCCAGAGGAGACUCCUUAUGAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCUUUGGGCGGCAAUUGCAUACCCUAGAA- -....---.((((-----((..((((((...(((....))).......(((((.....)))))((((((...((.......))....))))))))))))..))))))...- ( -38.90) >DroYak_CAF1 4385 102 - 1 -AUAU---AUGUA-----GUUAGCAGUUCCAGAGGAGACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUUGGGCGGCAAUUGCAUACCCUAGAAC -...(---(((((-----(((..........(((....))).......(((((.....)))))((((((...(((........))).)))))))))))))))......... ( -33.30) >consensus _AUAUACUAUGUAAUGU_GUUAGCA_UUCUG_____GACUCCUUAUAAAGAGCAAAGUGCUCUGCCGCCAGUGACACACAUUCGUGGAGCGGCAAUUGCAUACCCUAGAA_ ........(((((((.................................(((((.....)))))((((((..(((.......)))..).))))).))))))).......... (-17.16 = -18.64 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:06 2006