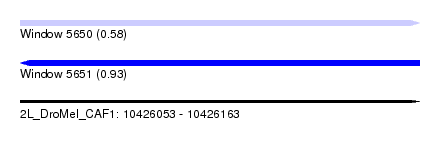
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,426,053 – 10,426,163 |
| Length | 110 |
| Max. P | 0.932768 |

| Location | 10,426,053 – 10,426,163 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
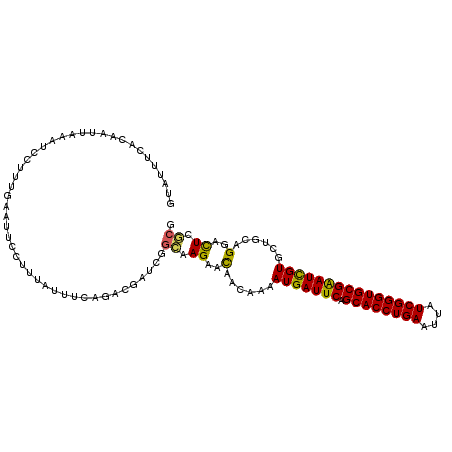
>2L_DroMel_CAF1 10426053 110 + 22407834 CGCGAGUCCUGCAGCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUGAAAUAAAGGAAUUCAAAGGAUUUAAUUGUGAAAUAC ((((((((((((((((.((((((((((.((....)).))))).)))))....)))))).......((...(((........)))..))..)))))....)))))...... ( -29.90) >DroSim_CAF1 2299 110 + 1 CGCGAAUCCUGCAGCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUGCAAAAAAGGAGUUCACAGGAUUAAAUUGUGAAAUAC ......((((((((((.((((((((((.((....)).))))).)))))....))))))..((((.(.....).))))...)))).(((((((.......))))))).... ( -32.50) >DroEre_CAF1 2315 110 + 1 CGCGAGUCCUGUAGCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUGAAAUAAAGGAGUUCAAGGGAUUUAAUCGUGAAAUAC ((((((((((..(((..((((((((((.((....)).))))).))))).(((((((..((..((.....))..)))))))))..)))...)))))....)))))...... ( -29.90) >DroYak_CAF1 2270 110 + 1 CGCGAGUCCUGAAGCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUUGUCUAAAACAAAGAAAUACAAAGGAUUUAAUUGUGAAAUAC ((((((((((.......((((((((((.((....)).))))).)))))...((((..(((((.................)))))..)))))))))....)))))...... ( -27.93) >DroMoj_CAF1 2652 94 + 1 CUUGAAUCCUGCAGAACAAUCCGCACCCGAUAGUUCAGGUGCUGAAUCAUUUUAUUAUUCUUGCCAAUAGUCUAAAAAAAAAAAAA--A--------------AAAUAGA ...((.....((((((......(((((.((....)).)))))((((....))))...))).)))......))..............--.--------------....... ( -12.60) >DroAna_CAF1 2216 92 + 1 CGAGAGUCCUGGAGAACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUUCCGAUAGUCUGAAAUAAA------------------UGGUGAAAUAU .(((((..(.(.((((.((((((((((.((....)).))))).))))).)))).).)..))))).................------------------........... ( -23.10) >consensus CGCGAGUCCUGCAGCACGAUUCGCACCCGAUAAUUCAGGUGCUGAAUCAUUUUGUUGUUCUUGCCGAUCGUCUGAAAUAAAGGAAUUCAAAGGAUUUAAUUGUGAAAUAC .(((((...........((((((((((.((....)).))))).)))))...........))))).............................................. (-16.59 = -17.03 + 0.45)

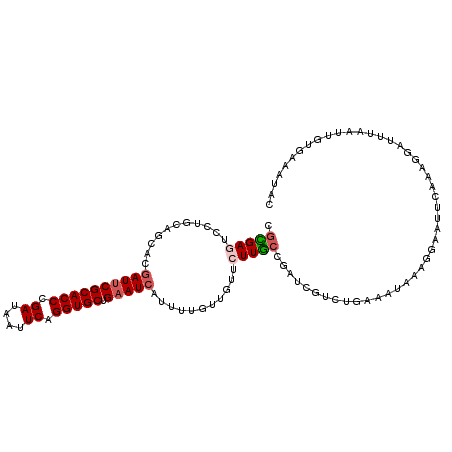
| Location | 10,426,053 – 10,426,163 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -21.36 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10426053 110 - 22407834 GUAUUUCACAAUUAAAUCCUUUGAAUUCCUUUAUUUCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCG ...............(((.((((((.........)))))).)))..((.((..(..((..(((((((.((((((((....)))))))))))))))..))..)..)).)). ( -30.10) >DroSim_CAF1 2299 110 - 1 GUAUUUCACAAUUUAAUCCUGUGAACUCCUUUUUUGCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGAUUCGCG ....................(((((.(((((((((((.(.....).)))))))...((..(((((((.((((((((....)))))))))))))))..)).))))))))). ( -36.00) >DroEre_CAF1 2315 110 - 1 GUAUUUCACGAUUAAAUCCCUUGAACUCCUUUAUUUCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUACAGGACUCGCG ........(((((........((((....))))........)))))((.((..(......(((((((.((((((((....)))))))))))))))......)..)).)). ( -27.19) >DroYak_CAF1 2270 110 - 1 GUAUUUCACAAUUAAAUCCUUUGUAUUUCUUUGUUUUAGACAAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUUCAGGACUCGCG ................((((((((..(((((.(((..........))))))))..)))))(((((((.((((((((....)))))))))))))))......)))...... ( -27.70) >DroMoj_CAF1 2652 94 - 1 UCUAUUU--------------U--UUUUUUUUUUUUUAGACUAUUGGCAAGAAUAAUAAAAUGAUUCAGCACCUGAACUAUCGGGUGCGGAUUGUUCUGCAGGAUUCAAG .......--------------.--......................(((.(((((((...........((((((((....))))))))..)))))))))).......... ( -20.52) >DroAna_CAF1 2216 92 - 1 AUAUUUCACCA------------------UUUAUUUCAGACUAUCGGAAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUUCUCCAGGACUCUCG ........((.------------------....((((.(.....).)))).........((((((((.((((((((....)))))))))))))))).....))....... ( -23.70) >consensus GUAUUUCACAAUUAAAUCCUUUGAAUUCCUUUAUUUCAGACGAUCGGCAAGAACAACAAAAUGAUUCAGCACCUGAAUUAUCGGGUGCGAAUCGUGCUGCAGGACUCGCG ..............................................((.((..(......(((((((.((((((((....)))))))))))))))......)..)).)). (-21.36 = -20.83 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:00 2006