| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,424,958 – 10,425,057 |
| Length | 99 |
| Max. P | 0.667136 |

| Location | 10,424,958 – 10,425,057 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.21 |
| Energy contribution | -17.85 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
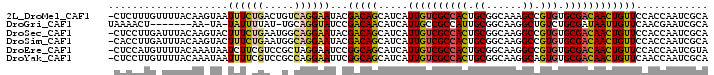
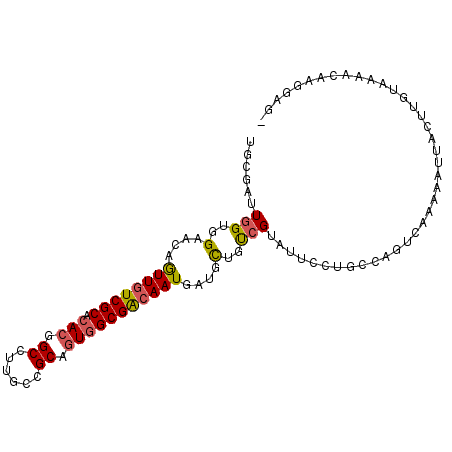
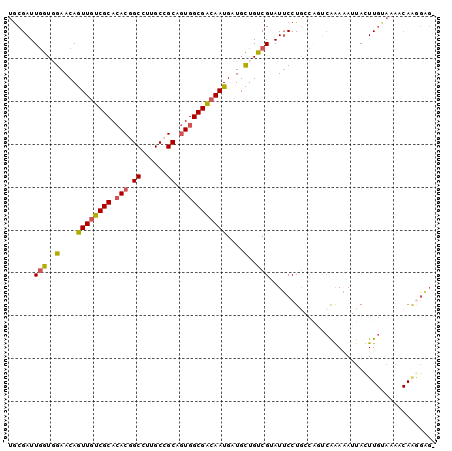
>2L_DroMel_CAF1 10424958 99 + 22407834 -CUCUUUGUUUUACAAGUAAUUUCUGACUGUCAGGAAUACGACAGCAUCAUUGUCGCCACUGCGGCAAAGCCGUGUGCGACAACUGUUCCACCAAUCGCA -...............(((.((((((.....)))))))))(((((.....(((((((.((.(((((...)))))))))))))))))))............ ( -27.60) >DroGri_CAF1 1553 90 + 1 UAAAACU-------AA-UA-UAUUUUAU-UGCAGGUAUCCGACAACAUCAUUGCCGCCAUUGCGGCAAGGCUGUCUGCGAUAAUUGUUCAACGAAUCGCA .......-------..-..-....((((-(((((..(.((((.....)).(((((((....))))))))).)..)))))))))................. ( -25.80) >DroSec_CAF1 1220 99 + 1 -CUCCUUGAUUUACAAGUACUUUCUGAAUGGCAGGAAUACGACAGCAUCAUUGUCGCCACUGCGGCAAGGCCGUGUGCGACAACUGUUCCACCAAUCGCA -...((((.....))))........((.(((..((((((.((.....)).(((((((.((.(((((...)))))))))))))).)))))).))).))... ( -28.30) >DroSim_CAF1 1205 99 + 1 -CACCUUGAUUUACAAGUACUUUCUGAAUGGCAGGAAUACGACAGCAUCAUUGUCGCCACUGCGGCAAGGCCGUGUGCGACAACUGUUCCACCAAUCGCA -...((((.....))))........((.(((..((((((.((.....)).(((((((.((.(((((...)))))))))))))).)))))).))).))... ( -28.30) >DroEre_CAF1 1224 99 + 1 -CUCCAUGUUUUACAAAUAAUCUUCGUCCGCUAGGAAUCCGGCAGCAUCAUUGUCGCCACUGCGGCAAGGCCGUGUGCGACAACUGUUCCACCAAUCGUA -..........(((............(((....)))....((.((((...(((((((.((.(((((...)))))))))))))).)))))).......))) ( -25.80) >DroYak_CAF1 1179 99 + 1 -CUCCUUGUUUUACAAAUAAUUUUCGUCCGCCAGGAAUUCGGCAGCAUCAUUGUCGCCACUGCGGCAAGGCAGUGUGCGACAACUGUUCAACCAAUCGCA -......(((..(((..............(((........))).......(((((((((((((......)))))).))))))).)))..)))........ ( -28.00) >consensus _CUCCUUGUUUUACAAGUAAUUUCUGACUGGCAGGAAUACGACAGCAUCAUUGUCGCCACUGCGGCAAGGCCGUGUGCGACAACUGUUCCACCAAUCGCA ....................((((((.....))))))...(((((.....((((((((((.((......)).))).))))))))))))............ (-18.21 = -17.85 + -0.36)
| Location | 10,424,958 – 10,425,057 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
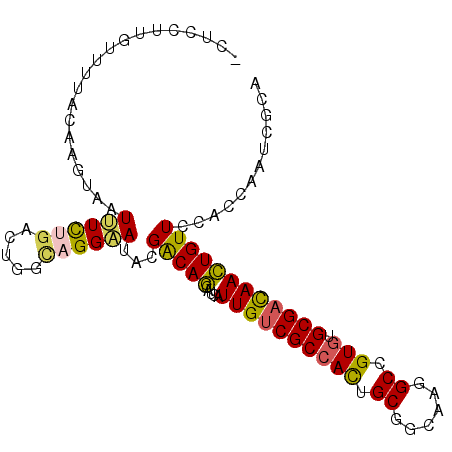
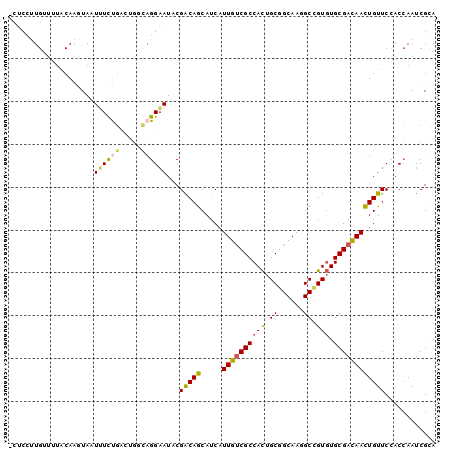
>2L_DroMel_CAF1 10424958 99 - 22407834 UGCGAUUGGUGGAACAGUUGUCGCACACGGCUUUGCCGCAGUGGCGACAAUGAUGCUGUCGUAUUCCUGACAGUCAGAAAUUACUUGUAAAACAAAGAG- (((((.((((......((((((((.(((.((......)).)))))))))))...(((((((......))))))).....)))).)))))..........- ( -29.10) >DroGri_CAF1 1553 90 - 1 UGCGAUUCGUUGAACAAUUAUCGCAGACAGCCUUGCCGCAAUGGCGGCAAUGAUGUUGUCGGAUACCUGCA-AUAAAAUA-UA-UU-------AGUUUUA ((((((..(((....))).))))))((((((.(((((((....)))))))....))))))...........-.((((((.-..-..-------.)))))) ( -25.70) >DroSec_CAF1 1220 99 - 1 UGCGAUUGGUGGAACAGUUGUCGCACACGGCCUUGCCGCAGUGGCGACAAUGAUGCUGUCGUAUUCCUGCCAUUCAGAAAGUACUUGUAAAUCAAGGAG- ...((.(((..(....((((((((.(((.((......)).)))))))))))(((((....))))).)..))).))........((((.....))))...- ( -31.70) >DroSim_CAF1 1205 99 - 1 UGCGAUUGGUGGAACAGUUGUCGCACACGGCCUUGCCGCAGUGGCGACAAUGAUGCUGUCGUAUUCCUGCCAUUCAGAAAGUACUUGUAAAUCAAGGUG- ...((.(((..(....((((((((.(((.((......)).)))))))))))(((((....))))).)..))).))........((((.....))))...- ( -31.70) >DroEre_CAF1 1224 99 - 1 UACGAUUGGUGGAACAGUUGUCGCACACGGCCUUGCCGCAGUGGCGACAAUGAUGCUGCCGGAUUCCUAGCGGACGAAGAUUAUUUGUAAAACAUGGAG- .....((((..(....((((((((.(((.((......)).)))))))))))....)..))))..(((......(((((.....))))).......))).- ( -31.82) >DroYak_CAF1 1179 99 - 1 UGCGAUUGGUUGAACAGUUGUCGCACACUGCCUUGCCGCAGUGGCGACAAUGAUGCUGCCGAAUUCCUGGCGGACGAAAAUUAUUUGUAAAACAAGGAG- (((((.(((((.....((((((((.((((((......))))))))))))))..((((((((......)))))).))..))))).)))))..........- ( -35.50) >consensus UGCGAUUGGUGGAACAGUUGUCGCACACGGCCUUGCCGCAGUGGCGACAAUGAUGCUGUCGUAUUCCUGCCAGUCAAAAAUUACUUGUAAAACAAGGAG_ ......(((..(....((((((((.(((.((......)).)))))))))))....)..)))....................................... (-19.59 = -19.62 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:56 2006