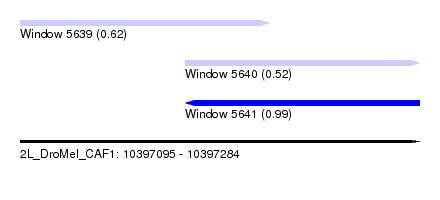
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,397,095 – 10,397,284 |
| Length | 189 |
| Max. P | 0.993612 |

| Location | 10,397,095 – 10,397,213 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10397095 118 + 22407834 GAUUUGCCAUAAUUAAUAAACAAUAAAUCACAGCUG--UGGUUUGGUAAAUAAGUGCAGCGUGAACAAUGUACGCUUAUGUACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUA ((((..(.(((................((((.((((--((.((........)).))))))))))...((((((((.(((((............))))).)))))))).))).)..)))). ( -29.70) >DroSec_CAF1 12046 120 + 1 GAUUUGCCAUAAUUAAUAAACAAUAAAUCACAGCUGUGUGGUUUGGUAAAUAAGUGCAGCGUGAAUAAUGUACGCUUAUGUACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUA ((((..(.(((................((((.((((..(...((....))...)..))))))))...((((((((.(((((............))))).)))))))).))).)..)))). ( -29.60) >DroSim_CAF1 11943 120 + 1 GAUUUGCCAUAAUUAAUAAACAAUAAAUCACAGCUGUGUGGUUUGGUAAAUAAGUGCAGCGUGAAUAAUGUACGCUUAUGUACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUA ((((..(.(((................((((.((((..(...((....))...)..))))))))...((((((((.(((((............))))).)))))))).))).)..)))). ( -29.60) >DroEre_CAF1 12642 110 + 1 GAUUUGUCAUUUUUAAUAAACAAUAAAUUGCAACUGUGUGGAUUGGUAAAUAAGUGCAGCGUGAACAUUA----------UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGGUUA ..................(((......(..(..(((..(..(((....)))..)..))).)..).(((((----------((.((..((((........))))..)).))))))).))). ( -18.20) >DroYak_CAF1 12683 119 + 1 GAUUUGCCACAAAUAAUAAACAAUAAAUUGCAAGUGUGUGGUUUGGUAAAUAAGUGCAGCGUGAACAAUGUACGC-UAUGUACAUAAACAUUCGCAUAUGUGUGCGUAUAUAGUGGGUUA .((((((((................(((..((....))..)))))))))))..(((((..(....)..)))))((-(((((((.((.((((......)))).)).)))))))))...... ( -27.79) >consensus GAUUUGCCAUAAUUAAUAAACAAUAAAUCACAGCUGUGUGGUUUGGUAAAUAAGUGCAGCGUGAACAAUGUACGCUUAUGUACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUA ......((((.............(((((((((....)))))))))(((.(((.((((.(.(((....(((((((....)))))))...))).))))).))).))).......)))).... (-20.94 = -21.48 + 0.54)



| Location | 10,397,173 – 10,397,284 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10397173 111 + 22407834 UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUAACAAAUUGCUGAUGUACACAUAUUAAGUGCUUUUACAAUGCAUACGCAAAUAUCGAUGCAAAUUCUAGUAA (((.....((((...((((((((((((...(((..(.........)..)))))))))))))))..)))).........(((((.((.......))))))).......))). ( -24.20) >DroSec_CAF1 12126 111 + 1 UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUAACAAAUUGCUGAUGUACAACUAUUAAGUGAUUUUACAAUGCAUACGCAAAUAUCGAUGCAUAUUCUAGUAA (((.....((((.((((....))))((((((((..(.........)..)).))))))........))))........((((((.((.......))))))))......))). ( -20.00) >DroSim_CAF1 12023 111 + 1 UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUAACAAAUUGCUGAUGUACAACUAUUAAGUGGUUUUACAAUGCAUACGCAAAUAUCGAUGUAAAUUCUAGUAA .......(((.(((.((((...(((((((...((......)).....((...((((.((((((...)))))).))))..))))))))).))))))))))............ ( -23.70) >DroEre_CAF1 12712 111 + 1 UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGGUUAACAAAUUACGGAUGUACAACUACUAGGUGAUUUUACAAUGCAUACGCAAAUAUCGAUGCAAAUUCUAUGAA ..((((..(((((((((....)))))....((((((....(((.........)))....)))))))))).........(((((.((.......))))))).....)))).. ( -20.60) >DroYak_CAF1 12762 111 + 1 UACAUAAACAUUCGCAUAUGUGUGCGUAUAUAGUGGGUUAACAAAUUGCUGAUGUACAACUACUAAGUGAUUUUACAAUGCAUACGCAAAUAUCGAUGCAAAUUCUAUGAA ..((((...(((.((((((((.(((((((.((....((....((((..((...(((....)))..))..)))).))..)).))))))).))))..)))).)))..)))).. ( -22.20) >consensus UACAUAAACACUCGCAUAUGUGUGCGUAUAUAGUGGAUUAACAAAUUGCUGAUGUACAACUAUUAAGUGAUUUUACAAUGCAUACGCAAAUAUCGAUGCAAAUUCUAGUAA .............((((((((.(((((.....(((.(((...((((((((...(((....)))..))))))))...))).)))))))).))))..))))............ (-18.60 = -18.08 + -0.52)


| Location | 10,397,173 – 10,397,284 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -21.31 |
| Energy contribution | -21.19 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10397173 111 - 22407834 UUACUAGAAUUUGCAUCGAUAUUUGCGUAUGCAUUGUAAAAGCACUUAAUAUGUGUACAUCAGCAAUUUGUUAAUCCACUAUAUACGCACACAUAUGCGAGUGUUUAUGUA ...........(((((.((((((((((((((...(((....((((.......)))).....(((.....)))..............)))..)))))))))))))).))))) ( -24.50) >DroSec_CAF1 12126 111 - 1 UUACUAGAAUAUGCAUCGAUAUUUGCGUAUGCAUUGUAAAAUCACUUAAUAGUUGUACAUCAGCAAUUUGUUAAUCCACUAUAUACGCACACAUAUGCGAGUGUUUAUGUA ...........(((((.((((((((((((((...((((.......(((((((((((......)))).))))))).........))))....)))))))))))))).))))) ( -24.29) >DroSim_CAF1 12023 111 - 1 UUACUAGAAUUUACAUCGAUAUUUGCGUAUGCAUUGUAAAACCACUUAAUAGUUGUACAUCAGCAAUUUGUUAAUCCACUAUAUACGCACACAUAUGCGAGUGUUUAUGUA ...........(((((.((((((((((((((...((((.......(((((((((((......)))).))))))).........))))....)))))))))))))).))))) ( -24.19) >DroEre_CAF1 12712 111 - 1 UUCAUAGAAUUUGCAUCGAUAUUUGCGUAUGCAUUGUAAAAUCACCUAGUAGUUGUACAUCCGUAAUUUGUUAACCCACUAUAUACGCACACAUAUGCGAGUGUUUAUGUA ...........(((((.((((((((((((((...(((.........((((.((((.(((.........)))))))..)))).....)))..)))))))))))))).))))) ( -22.84) >DroYak_CAF1 12762 111 - 1 UUCAUAGAAUUUGCAUCGAUAUUUGCGUAUGCAUUGUAAAAUCACUUAGUAGUUGUACAUCAGCAAUUUGUUAACCCACUAUAUACGCACACAUAUGCGAAUGUUUAUGUA ...........(((((.((((((((((((((...(((.........((((.((((.(((.........)))))))..)))).....)))..)))))))))))))).))))) ( -23.54) >consensus UUACUAGAAUUUGCAUCGAUAUUUGCGUAUGCAUUGUAAAAUCACUUAAUAGUUGUACAUCAGCAAUUUGUUAAUCCACUAUAUACGCACACAUAUGCGAGUGUUUAUGUA ...........(((((.((((((((((((((...((((.......(((((((((((......)))).))))))).........))))....)))))))))))))).))))) (-21.31 = -21.19 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:50 2006