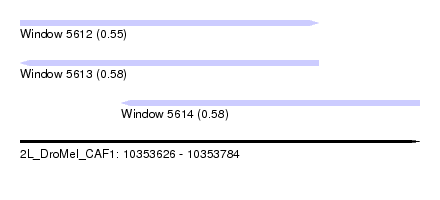
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,353,626 – 10,353,784 |
| Length | 158 |
| Max. P | 0.583184 |

| Location | 10,353,626 – 10,353,744 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -19.84 |
| Energy contribution | -21.09 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10353626 118 + 22407834 UCCAUUAUCCUUGGCUUGGGUAAACAAAUAAAUGCCGUUGCUUUUUU--AUGCGUUCGAGCAUAAGUUGCUGUAACGAAACUAAGCUGCCGACUAUUUUAUAGUCAACACCAAUGGCAUU ....((((((.......)))))).......(((((((((((((....--...((((..((((.....))))..)))).....))))....((((((...))))))......))))))))) ( -31.40) >DroSim_CAF1 6325 118 + 1 UCCAUUAUCCUUGGCUUGGGUAAACAAAUAAAUGCCGCUGCUUUUUU--AUGCGUUCGAGCAUAAGUUGCUGUAACGAAACUAAGCUGCCGACUAUUUUAUAGUCAACACCAAUGGCAAU ....((((((.......)))))).........((((((.((((....--...((((..((((.....))))..)))).....)))).)).((((((...)))))).........)))).. ( -30.60) >DroYak_CAF1 6341 118 + 1 UCCAUUAUCCUUGGCUUGGGUAAAUAAAUAAAUGCCGCUGCUUUUUU--AUGCGUUCGAGCAUAAGUUGCUGUAAGGAAACUAAGCUGCCGACUAUUUUAUAGUCAACACCAAUGGCAUU ....((((((.......)))))).......((((((((.((((.(((--((((......)))))))........((....)))))).)).((((((...)))))).........)))))) ( -32.30) >DroAna_CAF1 6014 118 + 1 UCCAUA--CUUUUGCUAAAAUAAAUAAAUAAUCGCGGCAGAUUUUUUCGCUGCGUUGGAGCAUAAGUUGCUGUAACGGAACUUAGUUGCCGACUAUUUUAUAGUCAACACCAAUAGCAUU ......--....(((((...............((((((.((.....))))))))((((((((.....))))(((((........))))).((((((...))))))....))))))))).. ( -30.90) >consensus UCCAUUAUCCUUGGCUUGGGUAAACAAAUAAAUGCCGCUGCUUUUUU__AUGCGUUCGAGCAUAAGUUGCUGUAACGAAACUAAGCUGCCGACUAUUUUAUAGUCAACACCAAUGGCAUU ....((((((.......))))))..........(((((.((((.........((((..((((.....))))..)))).....)))).)).((((((...)))))).........)))... (-19.84 = -21.09 + 1.25)

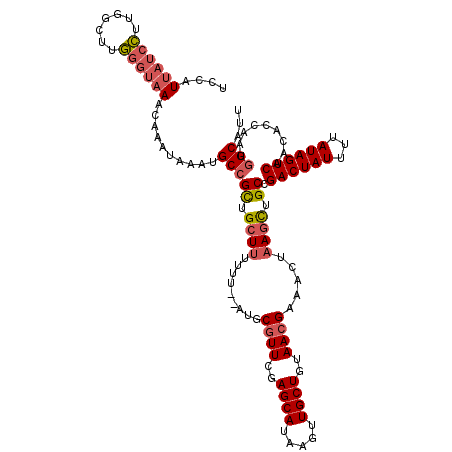

| Location | 10,353,626 – 10,353,744 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10353626 118 - 22407834 AAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAGCAACGGCAUUUAUUUGUUUACCCAAGCCAAGGAUAAUGGA ((((((.....((((((((((...))))))))))((((..((.((((..((((.....))))..))))...--))..))))...))))))...........(((..(....)....))). ( -29.30) >DroSim_CAF1 6325 118 - 1 AUUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAGCAGCGGCAUUUAUUUGUUUACCCAAGCCAAGGAUAAUGGA ....((((((.((((((((((...)))))))))).(((.((((.(.((.(((((...((((.((...((..--......))..))))))....))))))).).)))).)))..)))))). ( -28.50) >DroYak_CAF1 6341 118 - 1 AAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCCUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAGCAGCGGCAUUUAUUUAUUUACCCAAGCCAAGGAUAAUGGA ....((((((.((((((((((...)))))))))).)......(((((...((...((((((......))))--))....))...(((..................)))))))).))))). ( -30.77) >DroAna_CAF1 6014 118 - 1 AAUGCUAUUGGUGUUGACUAUAAAAUAGUCGGCAACUAAGUUCCGUUACAGCAACUUAUGCUCCAACGCAGCGAAAAAAUCUGCCGCGAUUAUUUAUUUAUUUUAGCAAAAG--UAUGGA ..(((((((((((((((((((...))))))))))..((((((.(......).))))))....))))(((.(((........))).)))...............)))))....--...... ( -27.80) >consensus AAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU__AAAAAAGCAGCGGCAUUUAUUUAUUUACCCAAGCCAAGGAUAAUGGA ..((((...(.((((((((((...)))))))))).).......((((..((((.....))))..))))................))))................................ (-20.24 = -20.55 + 0.31)

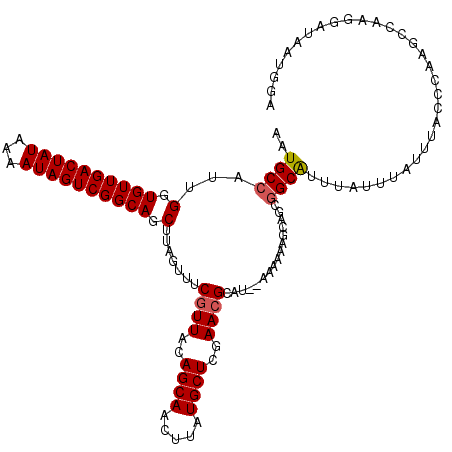
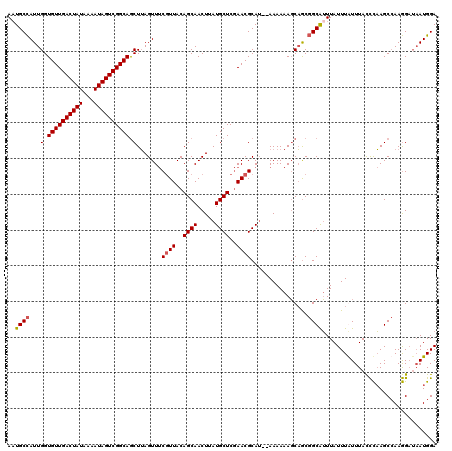
| Location | 10,353,666 – 10,353,784 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10353666 118 - 22407834 CGCAGCAAUAAAGAGCCAACUGAGGUGAUUAGGCACGCCGAAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAG .((.((........))....(((.(.((((((((..(((((......)))))(((((((((...))))))))).)))))))).).)))..))...((((((......))))--))..... ( -31.40) >DroSim_CAF1 6365 118 - 1 CGCAGCAAUAAAGAGCCAACUGAGGUGAUUAGGCACGCCGAUUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAG .((.((........)).((((((((..(((.((....)))))..))...(.((((((((((...)))))))))).)))))))........))...((((((......))))--))..... ( -32.20) >DroYak_CAF1 6381 118 - 1 CGCAGCAAUAAAGAGCCAACUGAGGUGAUUAGGCACGCCGAAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCCUUACAGCAACUUAUGCUCGAACGCAU--AAAAAAG .((.((........))....(((((.((((((((..(((((......)))))(((((((((...))))))))).)))))))).)))))..))...((((((......))))--))..... ( -35.80) >DroAna_CAF1 6052 120 - 1 CGCAGCGAUAAAGAGCCAAGUGAAGUGAUUAGGCACGCCGAAUGCUAUUGGUGUUGACUAUAAAAUAGUCGGCAACUAAGUUCCGUUACAGCAACUUAUGCUCCAACGCAGCGAAAAAAU (((.(((.....((((.(((((..(((((..((.(((((((......)))))(((((((((...)))))))))......)).)))))))..).))))..))))...))).)))....... ( -33.50) >consensus CGCAGCAAUAAAGAGCCAACUGAGGUGAUUAGGCACGCCGAAUGCCAUUGGUGUUGACUAUAAAAUAGUCGGCAGCUUAGUUUCGUUACAGCAACUUAUGCUCGAACGCAU__AAAAAAG ....((......((((....((..(((((.((((..(((((......)))))(((((((((...)))))))))......)))).)))))..))......))))....))........... (-26.12 = -26.62 + 0.50)

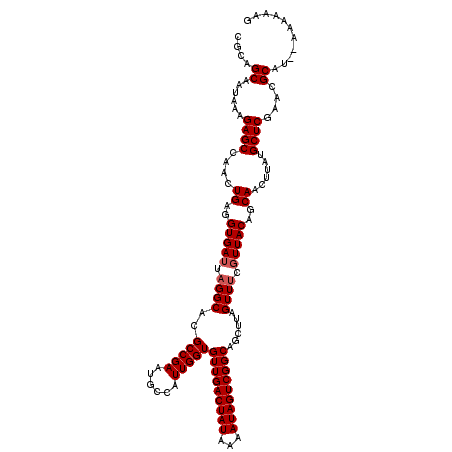
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:22 2006