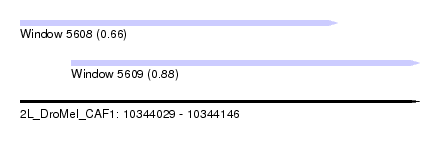
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,344,029 – 10,344,146 |
| Length | 117 |
| Max. P | 0.876761 |

| Location | 10,344,029 – 10,344,122 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -14.61 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10344029 93 + 22407834 GUUAGCCUUUAGUUA--------------------AGUGGUGGUAAUUAAUCAUUCAACUUAUGAGGAUUAUUUCUUAAAGGCCU-CUGGCUCGGAAAUAGUCUGAACCUUAUU ....((((((((((.--------------------((((((........)))))).))))...(((((....)))))))))))..-..((.(((((.....))))).))..... ( -22.00) >DroVir_CAF1 2110 113 + 1 GUUUCUCUUUAAAAAAAAAAUGUUAAUUUAGUUUUUGUGAAGCGAAUUGACUAUUCAACUUAUGAUUACUAUUUCAUAAAGGCUU-CUGGCUGGGAAAUAUUAUGAACGGUUUU (((((((..((((((..((((....))))..)))))).(((((((((.....))))...((((((........))))))..))))-).....)))))))............... ( -20.50) >DroSec_CAF1 679 93 + 1 GUUAGCCUGUAGAUA--------------------AGUGGUGGUAAUUAAUUAUUCAACUUAUGAGGAUUAUUUCUUAAAGGCCU-CUGGCUCGGAAAUAGUCUGAACUUUAUU (((.........(((--------------------(((.(((((.....)))))...))))))..(((((((((((....((((.-..)))).))))))))))).)))...... ( -18.80) >DroSim_CAF1 626 93 + 1 GUUAGCCUGUAGAUA--------------------AGUGGUGGUAAUUAAUUAUUCAACUUAUGAAGAUUAUUUCUUAAAGGCCU-CUGGCUCGGAAAUGGUCUGAACUUUGUU (((.........(((--------------------(((.(((((.....)))))...))))))..(((((((((((....((((.-..)))).))))))))))).)))...... ( -18.40) >DroEre_CAF1 629 90 + 1 ----GCCUUUAGUUA--------------------GGUGGUGCAAAUUAAUUAUUCAACCUAUGAGGAUUAUUUCAUAAAGGCCUUCUGGCUCGGAAAUAGUCUGAAAUUUAUU ----((((......)--------------------)))(((...(((.....)))..))).....((((((((((.....((((....))))..)))))))))).......... ( -20.30) >DroYak_CAF1 625 92 + 1 GUUAGCCUUUAGUUA--------------------AGUGAUACAAAUUAAUUAUUCAACUUAUGAGGACUAUUUCUUAAAGGCCU--UGGCUCGGAAAUAGUCUGAACAUUAUU (((...(((.((((.--------------------((((((........)))))).))))...)))((((((((((....(((..--..))).))))))))))..)))...... ( -21.60) >consensus GUUAGCCUUUAGAUA____________________AGUGGUGCUAAUUAAUUAUUCAACUUAUGAGGAUUAUUUCUUAAAGGCCU_CUGGCUCGGAAAUAGUCUGAACUUUAUU .................................................................(((((((((((....((((....)))).))))))))))).......... (-14.61 = -14.50 + -0.11)

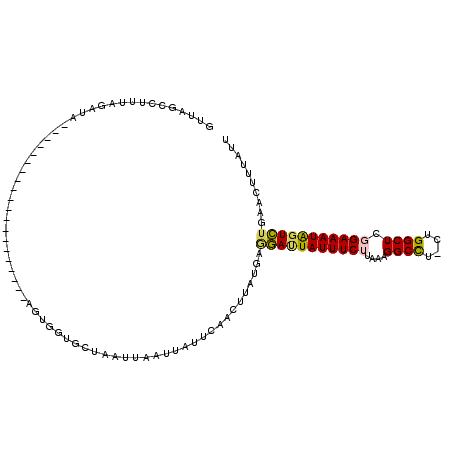
| Location | 10,344,044 – 10,344,146 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.27 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -15.83 |
| Energy contribution | -15.34 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10344044 102 + 22407834 -AGUGGUGGUAAUUAAUCAUUCAACUUAUGAGGAUUAUUUCUUAAAGGCCU-CUGGCUCGGAAAUAGUCUGAACCUUAUUGUA----CACGCG---------AUAAUUUUGAUGAUU -.............((((((.(((.......(((((((((((....((((.-..)))).))))))))))).....(((((((.----...)))---------))))..))))))))) ( -22.00) >DroVir_CAF1 2144 116 + 1 UUGUGAAGCGAAUUGACUAUUCAACUUAUGAUUACUAUUUCAUAAAGGCUU-CUGGCUGGGAAAUAUUAUGAACGGUUUUAUGUGUAAAUAUAAAAUGUUUGAUAUUUAGUUUGAUU ....(((((((((.....))))...((((((........))))))..))))-).(((((...((((((..(((((.((((((((....))))))))))))))))))))))))..... ( -22.70) >DroSec_CAF1 694 111 + 1 -AGUGGUGGUAAUUAAUUAUUCAACUUAUGAGGAUUAUUUCUUAAAGGCCU-CUGGCUCGGAAAUAGUCUGAACUUUAUUGUA----CACGCGAGGUUUGCAAUAAUUUUGAUGAUU -.............((((((.(((.......(((((((((((....((((.-..)))).))))))))))).....((((((((----..(....)...))))))))..))))))))) ( -23.70) >DroEre_CAF1 640 112 + 1 -GGUGGUGCAAAUUAAUUAUUCAACCUAUGAGGAUUAUUUCAUAAAGGCCUUCUGGCUCGGAAAUAGUCUGAAAUUUAUUGUA----CAUGCGAGGUGUGCAAUAAUUUUGAUGAUU -.............(((((((..........((((((((((.....((((....))))..)))))))))).(((.((((((((----(((.....))))))))))).)))))))))) ( -28.50) >DroYak_CAF1 640 110 + 1 -AGUGAUACAAAUUAAUUAUUCAACUUAUGAGGACUAUUUCUUAAAGGCCU--UGGCUCGGAAAUAGUCUGAACAUUAUUGUA----CAUGAGAGGUGUGCAAUAAUUUUGAUAAUU -.............((((((.(((.......(((((((((((....(((..--..))).)))))))))))....(((((((((----(((.....)))))))))))).))))))))) ( -29.50) >DroAna_CAF1 646 109 + 1 -UUUGGAAUAUAUUAAUCAUUCAUCUUAUGAUGAUUAUUUUAUAAAGGCU---UAGCCAGGAAAUAAUCUGAAAUUUAUUGUA----CGCUUGGACUGUAUAAUAAUCAUGAUGAUU -...................(((((..((((((((((((((.....(((.---..)))..))))))))).......(((((((----((.......))))))))))))))))))).. ( -21.80) >consensus _AGUGGUGCAAAUUAAUUAUUCAACUUAUGAGGAUUAUUUCAUAAAGGCCU_CUGGCUCGGAAAUAGUCUGAACUUUAUUGUA____CACGCGAGGUGUGCAAUAAUUUUGAUGAUU ..............(((((((..........((((((((((.....((((....))))..)))))))))).....((((((((...............))))))))....))))))) (-15.83 = -15.34 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:17 2006