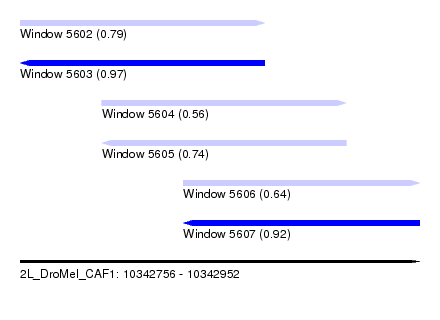
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,342,756 – 10,342,952 |
| Length | 196 |
| Max. P | 0.967616 |

| Location | 10,342,756 – 10,342,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
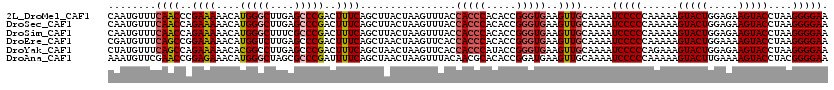
>2L_DroMel_CAF1 10342756 120 + 22407834 CAAUGUUUCAACCCGAAAAACAUGGGCUUGAGCCCGACUUUCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAA ..((((((.........))))))((((....))))((.(((((((((.....(....)(((((.....))))))))))).))).))((((.....(((((.....)))))....)))).. ( -33.20) >DroSec_CAF1 4359 120 + 1 CAAUGUUUCAACCAGAAAAACAUGGGCUUGAGCCCGACUUUCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAA ..((((((.........))))))((((....))))((.(((((((((.....(....)(((((.....))))))))))).))).))((((.....(((((.....)))))....)))).. ( -33.20) >DroSim_CAF1 2597 120 + 1 CAAUGUUUCAACCAGAAAAACAUGGGCUUUCGCCCGACUUUCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAA ..((((((.........))))))((((....))))((.(((((((((.....(....)(((((.....))))))))))).))).))((((.....(((((.....)))))....)))).. ( -32.90) >DroEre_CAF1 4411 120 + 1 CGAUGUUUCAGCCGGAAAAACAUGGUCUUGAGCCCGACUUUCAGCUAACUAAGUUCACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAAAAGUACCUAAGGGGAA ....((((.(((((........))).)).)))).((((((..((((.....))))...(((((.....))))))))))).....(((((......(((((.....)))))....))))). ( -30.40) >DroYak_CAF1 3234 120 + 1 CUAUGUUUCAGCCAGAAAAACACGGCCUUGAGCCCGACUUUCAGCUAACUAAGUUCACCACCCAUACCGGGUGAAGUUGCAAAAUCCCCCAGAAAGUACUGGAGAAGUACCUAAGGGGAA ...(((..((((..((((.....(((.....)))....)))).)))............(((((.....)))))..)..)))...(((((......(((((.....)))))....))))). ( -30.00) >DroAna_CAF1 4442 120 + 1 AAAUGUUCGAACCGGAGAAACAUGGGCUAGCGCCCGAUUUUCAGCUAACUAAGUUUACAACGCACACCGGAUGAAGUUGCAAAAUCCCCCAAAAAGUACUUGAAAAGUACCUACGGGGAA .....((((..((((.((((..(((((....)))))..))))((((.....))))...........)))).)))).........(((((......(((((.....)))))....))))). ( -30.10) >consensus CAAUGUUUCAACCAGAAAAACAUGGGCUUGAGCCCGACUUUCAGCUAACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAA ........((((..((((....(((((....)))))..))))................(((((.....)))))..)))).....(((((......(((((.....)))))....))))). (-27.08 = -27.58 + 0.50)

| Location | 10,342,756 – 10,342,876 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10342756 120 - 22407834 UUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGAAAGUCGGGCUCAAGCCCAUGUUUUUCGGGUUGAAACAUUG .((((((.(((((((.....)))))))...)))))).....((((.((((((.....)))))).............(((((((..((((....))))....)))))))))))........ ( -41.20) >DroSec_CAF1 4359 120 - 1 UUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGAAAGUCGGGCUCAAGCCCAUGUUUUUCUGGUUGAAACAUUG .((((((.(((((((.....)))))))...)))))).....(((((((((((.....)))))).((((((((....)))).....((((....))))..))))....)))))........ ( -38.90) >DroSim_CAF1 2597 120 - 1 UUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGAAAGUCGGGCGAAAGCCCAUGUUUUUCUGGUUGAAACAUUG .((((((.(((((((.....)))))))...)))))).....(((((((((((.....)))))).((((((((....)))).....((((....))))..))))....)))))........ ( -42.40) >DroEre_CAF1 4411 120 - 1 UUCCCCUUAGGUACUUUUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUGAACUUAGUUAGCUGAAAGUCGGGCUCAAGACCAUGUUUUUCCGGCUGAAACAUCG .((((((.(((((((.....)))))))...))))))(((.((((((((((((.....)))))).......)))).)).)))(((((((...(((((...))))))))))))......... ( -36.81) >DroYak_CAF1 3234 120 - 1 UUCCCCUUAGGUACUUCUCCAGUACUUUCUGGGGGAUUUUGCAACUUCACCCGGUAUGGGUGGUGAACUUAGUUAGCUGAAAGUCGGGCUCAAGGCCGUGUUUUUCUGGCUGAAACAUAG .((((((.(((((((.....)))))))...))))))...........(((((.....)))))(((...((((((((..(((...(((.(....).)))..)))..))))))))..))).. ( -40.20) >DroAna_CAF1 4442 120 - 1 UUCCCCGUAGGUACUUUUCAAGUACUUUUUGGGGGAUUUUGCAACUUCAUCCGGUGUGCGUUGUAAACUUAGUUAGCUGAAAAUCGGGCGCUAGCCCAUGUUUCUCCGGUUCGAACAUUU (((((((.(((((((.....)))))))..))))))).........(((..((((.(.((((.......((((....)))).....((((....))))))))..).))))...)))..... ( -36.40) >consensus UUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGAAAGUCGGGCUCAAGCCCAUGUUUUUCUGGUUGAAACAUUG ..((((..(((((((.....)))))))...))))((((((......((((((.....))))))......(((....)))))))))((((....))))(((((((.......))))))).. (-34.50 = -34.78 + 0.28)
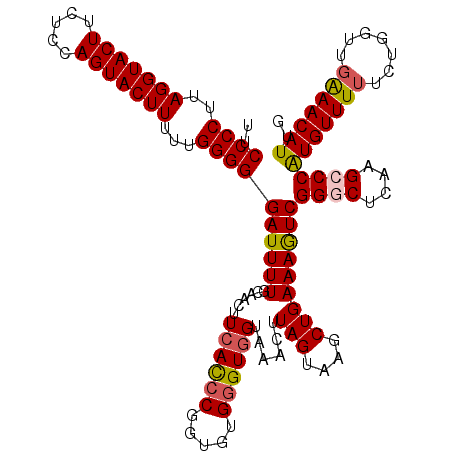
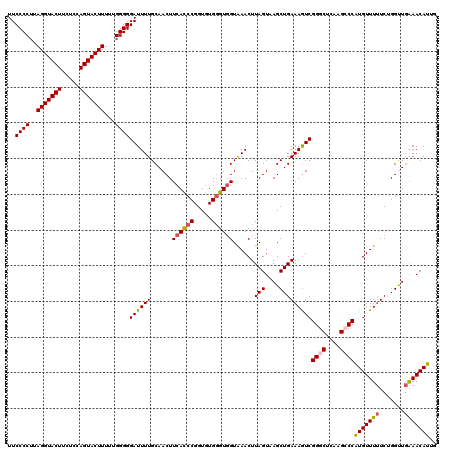
| Location | 10,342,796 – 10,342,916 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
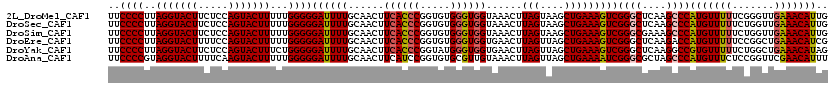
>2L_DroMel_CAF1 10342796 120 + 22407834 UCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAAUAAUGAUGGAUACCUUAUCGG .((((((.....(....)(((((.....))))))))))).....(((((......(((((.....)))))....))))).......................((((((.....)))))). ( -25.10) >DroSec_CAF1 4399 120 + 1 UCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAACGAUGGAUACCUUAUCGG ............((((..(((((.....)))))...........(((((......(((((.....)))))....))))).................))))..((((((.....)))))). ( -27.50) >DroSim_CAF1 2637 120 + 1 UCAGCUUACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAAUGAUGGAUACCUUAUCGG ............((((..(((((.....)))))...........(((((......(((((.....)))))....))))).................))))..((((((.....)))))). ( -25.30) >DroEre_CAF1 4451 114 + 1 UCAGCUAACUAAGUUCACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAAAAGUACCUAAGGGGAACAG------AAAAUAAGAACAAUGAUGGAUACCUUAUCGG ............((((..(((((.....)))))...........(((((......(((((.....)))))....)))))....------.......))))..((((((.....)))))). ( -27.80) >DroYak_CAF1 3274 120 + 1 UCAGCUAACUAAGUUCACCACCCAUACCGGGUGAAGUUGCAAAAUCCCCCAGAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAAAAAUGAUGGAUACCUUAUCGG ........(((..(((..(((((.....)))))...........(((((......(((((.....)))))....)))))...))).))).............((((((.....)))))). ( -25.60) >DroAna_CAF1 4482 120 + 1 UCAGCUAACUAAGUUUACAACGCACACCGGAUGAAGUUGCAAAAUCCCCCAAAAAGUACUUGAAAAGUACCUACGGGGAACAGACAUAGCUAAUACAAACAAUGAAGGAUAUCUUAUUGG ..(((((..............(((.((........)))))....(((((......(((((.....)))))....))))).......)))))........((((((........)))))). ( -22.60) >consensus UCAGCUAACUAAGUUUACCACCCACACCGGGUGAAGUUGCAAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAAUGAUGGAUACCUUAUCGG ............((((..(((((.....)))))...........(((((......(((((.....)))))....))))).................))))..((((((.....)))))). (-22.08 = -22.17 + 0.09)
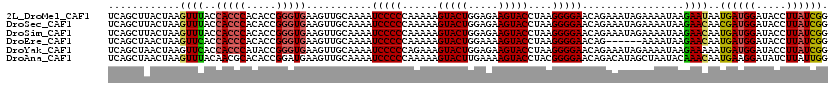


| Location | 10,342,796 – 10,342,916 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10342796 120 - 22407834 CCGAUAAGGUAUCCAUCAUUAUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGA ..(((((((((........)).)))))))..........(((((((..(((((((.....)))))))...))))))).........((((((.....)))))).....((((....)))) ( -28.60) >DroSec_CAF1 4399 120 - 1 CCGAUAAGGUAUCCAUCGUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGA ....((((....((((((((((.................(((((((..(((((((.....)))))))...)))))))...)))))..(((.....))))))))....))))......... ( -31.15) >DroSim_CAF1 2637 120 - 1 CCGAUAAGGUAUCCAUCAUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGA ..(((((((.............)))))))..........(((((((..(((((((.....)))))))...))))))).........((((((.....)))))).....((((....)))) ( -28.52) >DroEre_CAF1 4451 114 - 1 CCGAUAAGGUAUCCAUCAUUGUUCUUAUUUU------CUGUUCCCCUUAGGUACUUUUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUGAACUUAGUUAGCUGA ..(((((((.............)))))))..------..(((((((..(((((((.....)))))))...)))))))...((((((((((((.....)))))).......)))).))... ( -30.13) >DroYak_CAF1 3274 120 - 1 CCGAUAAGGUAUCCAUCAUUUUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUCUGGGGGAUUUUGCAACUUCACCCGGUAUGGGUGGUGAACUUAGUUAGCUGA ..(((((((.............)))))))..........(((((((..(((((((.....)))))))...)))))))...((((((((((((.....)))))).......)))).))... ( -30.13) >DroAna_CAF1 4482 120 - 1 CCAAUAAGAUAUCCUUCAUUGUUUGUAUUAGCUAUGUCUGUUCCCCGUAGGUACUUUUCAAGUACUUUUUGGGGGAUUUUGCAACUUCAUCCGGUGUGCGUUGUAAACUUAGUUAGCUGA .(((((((.....))).))))......(((((((...((((((((((.(((((((.....)))))))..))))))).((((((((..(((.....))).))))))))..))).))))))) ( -32.50) >consensus CCGAUAAGGUAUCCAUCAUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUUGCAACUUCACCCGGUGUGGGUGGUAAACUUAGUAAGCUGA ..(((((((.............)))))))..........(((((((..(((((((.....)))))))...))))))).........((((((.....)))))).....((((....)))) (-26.16 = -26.10 + -0.05)

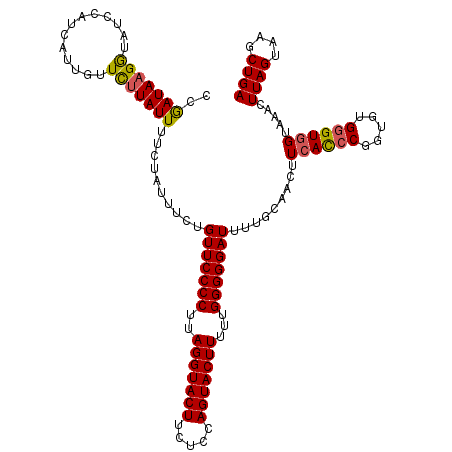
| Location | 10,342,836 – 10,342,952 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10342836 116 + 22407834 AAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAAUAAUGAUGGAUACCUUAUCGGUGAGAGAUAUUACUUUGUAUAAUCAGCAG----AUCUCGA ....(((((......(((((.....)))))....)))))..............................((((.....)))).(((((......(((......)))...----))))).. ( -20.40) >DroSec_CAF1 4439 109 + 1 AAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAACGAUGGAUACCUUAUCGGUGAGAGAUAGUACU-------AUCAGCAG----AUCUGGG ....(((((......(((((.....)))))....))))).((((...............((.((((((.....)))))).))...((((....)-------))).....----.)))).. ( -24.60) >DroSim_CAF1 2677 109 + 1 AAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAAUGAUGGAUACCUUAUCGGUGAGAGAUAGUACU-------AUCAGCAG----AUCUGGG ....(((((......(((((.....)))))....))))).((((..((.....)).......((((((.(((..((((.......)))))))))-------))))....----.)))).. ( -23.80) >DroEre_CAF1 4491 102 + 1 AAAAUCCCCCAAAAAGUACUGGAAAAGUACCUAAGGGGAACAG------AAAAUAAGAACAAUGAUGGAUACCUUAUCGGUGAGAGAUAGUGUU-------UUCAUCA-----CUAUCGG ....(((((......(((((.....)))))....)))))....------.............(((((((.((.(((((.......))))).)).-------)))))))-----....... ( -25.10) >DroYak_CAF1 3314 109 + 1 AAAAUCCCCCAGAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAAAAAUGAUGGAUACCUUAUCGGUGAGAGAUAGUACU-------AUCUGCAG----AUCUUGG ....(((((......(((((.....)))))....)))))..............(((((.....(((((.(((..((((.......)))))))))-------))).....----.))))). ( -23.20) >DroAna_CAF1 4522 113 + 1 AAAAUCCCCCAAAAAGUACUUGAAAAGUACCUACGGGGAACAGACAUAGCUAAUACAAACAAUGAAGGAUAUCUUAUUGGUAAGAAAUAAUGAU-------UUAUUUAUGUUAUGUUCAA ....(((((......(((((.....)))))....)))))...((((((((.........((((((........)))))).....((((((....-------))))))..))))))))... ( -23.80) >consensus AAAAUCCCCCAAAAAGUACUGGAGAAGUACCUAAGGGGAACAGAAAUAGAAAAUAAGAACAAUGAUGGAUACCUUAUCGGUGAGAGAUAGUACU_______AUCAGCAG____AUCUCGG ....(((((......(((((.....)))))....)))))..............................((((.....))))...................................... (-15.71 = -15.43 + -0.28)



| Location | 10,342,836 – 10,342,952 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10342836 116 - 22407834 UCGAGAU----CUGCUGAUUAUACAAAGUAAUAUCUCUCACCGAUAAGGUAUCCAUCAUUAUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUU ..(((((----.((((..........))))..)))))..(((.....))).............................(((((((..(((((((.....)))))))...)))))))... ( -26.20) >DroSec_CAF1 4439 109 - 1 CCCAGAU----CUGCUGAU-------AGUACUAUCUCUCACCGAUAAGGUAUCCAUCGUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUU ..((((.----.....(((-------(....))))....(((.....)))..........................)))).((((((.(((((((.....)))))))...)))))).... ( -24.40) >DroSim_CAF1 2677 109 - 1 CCCAGAU----CUGCUGAU-------AGUACUAUCUCUCACCGAUAAGGUAUCCAUCAUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUU ..((((.----.....(((-------(....))))....(((.....)))..........................)))).((((((.(((((((.....)))))))...)))))).... ( -24.40) >DroEre_CAF1 4491 102 - 1 CCGAUAG-----UGAUGAA-------AACACUAUCUCUCACCGAUAAGGUAUCCAUCAUUGUUCUUAUUUU------CUGUUCCCCUUAGGUACUUUUCCAGUACUUUUUGGGGGAUUUU ..(((((-----(((((..-------.((..((((.......))))..))...))))))))))........------..(((((((..(((((((.....)))))))...)))))))... ( -29.50) >DroYak_CAF1 3314 109 - 1 CCAAGAU----CUGCAGAU-------AGUACUAUCUCUCACCGAUAAGGUAUCCAUCAUUUUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUCUGGGGGAUUUU ..((((.----....((((-------(....)))))...(((.....)))............)))).............(((((((..(((((((.....)))))))...)))))))... ( -23.90) >DroAna_CAF1 4522 113 - 1 UUGAACAUAACAUAAAUAA-------AUCAUUAUUUCUUACCAAUAAGAUAUCCUUCAUUGUUUGUAUUAGCUAUGUCUGUUCCCCGUAGGUACUUUUCAAGUACUUUUUGGGGGAUUUU ....(((((.(((((((((-------.........(((((....))))).........))))))))....).)))))..((((((((.(((((((.....)))))))..))))))))... ( -24.07) >consensus CCGAGAU____CUGCUGAU_______AGUACUAUCUCUCACCGAUAAGGUAUCCAUCAUUGUUCUUAUUUUCUAUUUCUGUUCCCCUUAGGUACUUCUCCAGUACUUUUUGGGGGAUUUU ..........................................(((((((.............)))))))..........(((((((..(((((((.....)))))))...)))))))... (-19.39 = -19.14 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:15 2006