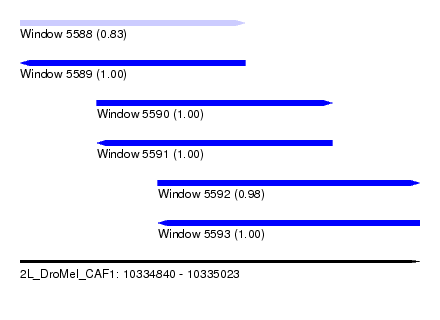
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,334,840 – 10,335,023 |
| Length | 183 |
| Max. P | 0.999901 |

| Location | 10,334,840 – 10,334,943 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334840 103 + 22407834 CAAAGAAACGAAUUUACAUUAAUUACGCACAUACACAAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACA ......................(((((((((......((....))......)))))))))((.((...(((((.(((....))).)))))...)).))..... ( -21.30) >DroSec_CAF1 701 103 + 1 CAAAGAAACGAAUUUACAUUAAUUACGCACAUACACCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCUCGCACACA ..((((....((((......))))..((..........))....))))....((((((((((......(((((.(((....))).)))))..)))))))))). ( -23.30) >DroSim_CAF1 701 103 + 1 CAAAGAAACGAAUUUACAUUAAUUACGCACAUACACCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACA ......................(((((((((......((....))......)))))))))((.((...(((((.(((....))).)))))...)).))..... ( -21.30) >DroEre_CAF1 392 103 + 1 CAAAGAAACGAAUUUACAUUAAUUACGCACACACACCAGCGUGCGCAUACCUGUGUGUGUGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACA ...............((((......(((((.(......).))))).......))))((((((.((...(((((.(((....))).)))))...)).)))))). ( -28.92) >DroYak_CAF1 686 103 + 1 CAAAGAAACGAAUUUACAUUAAUUACGCACACACACAAGCGCGCACAUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACA ..........................((.........(((.(((((((....))))))).)))((...(((((.(((....))).)))))...)).))..... ( -21.70) >consensus CAAAGAAACGAAUUUACAUUAAUUACGCACAUACACCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACA ........((((((......)))).))((((((((................)))))))).((.((...(((((.(((....))).)))))...)).))..... (-19.55 = -19.15 + -0.40)

| Location | 10,334,840 – 10,334,943 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -27.52 |
| Energy contribution | -26.96 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334840 103 - 22407834 UGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUUGUGUAUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG .(((.(..((...(((((((((....)))))))))...))..).)))(((((((((......)))))))))...(((((.....))))).............. ( -29.30) >DroSec_CAF1 701 103 - 1 UGUGUGCGAGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGGUGUAUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG ((((((.(((...(((((((((....))))))))).......))).))))))...((((((((((....))))))(((.((((......))))))).)))).. ( -29.00) >DroSim_CAF1 701 103 - 1 UGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGGUGUAUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG ((((((.(((...(((((((((....))))))))).......))).))))))...((((((((((....))))))(((.((((......))))))).)))).. ( -28.10) >DroEre_CAF1 392 103 - 1 UGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACACACACACAGGUAUGCGCACGCUGGUGUGUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG ((((((..((...(((((((((....)))))))))...))..)))))).(((..(((((((((((....))))))))))).....)))............... ( -39.70) >DroYak_CAF1 686 103 - 1 UGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAUGUGCGCGCUUGUGUGUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG .(((((..((...(((((((((....)))))))))...))....)))))(((..((((..(((((....)))))..)))).....)))............... ( -33.90) >consensus UGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGGUGUAUGUGCGUAAUUAAUGUAAAUUCGUUUCUUUG ((((((.(((...(((((((((....))))))))).......))).)))))).......((((((....))))))(((.((((......)))))))....... (-27.52 = -26.96 + -0.56)


| Location | 10,334,875 – 10,334,983 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -36.22 |
| Energy contribution | -35.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334875 108 + 22407834 CAAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACUUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGU ...(((((((.......(((((((....((...(((((.(((....))).)))))...))..)))))))((((((....((((....)))).)))))).))))))).. ( -39.90) >DroSec_CAF1 736 108 + 1 CCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCUCGCACACACGCGCACAUUUGCGAGUACUUGCCGUGCGCUGGUGUGUGU ...(((((((......(((((((((((......(((((.(((....))).)))))..))))))))))).((((((....((((....)))).)))))).))))))).. ( -42.20) >DroSim_CAF1 736 108 + 1 CCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACAUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGU (((((............(((((((.((.((...(((((.(((....))).)))))...)).)).)))))))((((....((((....)))).)))))))))....... ( -40.00) >DroEre_CAF1 427 108 + 1 CCAGCGUGCGCAUACCUGUGUGUGUGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACAAGCGCACUUUUGCGAGUGCUUGCCGUGCGGUGGUAUGUGU ((((((..(((((....)))))..))))))...(((((.(((....))).)))))((((((((((.(((((((.(......).)))))))..)))))..))))).... ( -45.50) >DroYak_CAF1 721 108 + 1 CAAGCGCGCACAUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACAAGCACACUUCUGCGAGUGCUUGCCGUGCGCUAGUGUGUGU ...((((((((.....((((((((....((...(((((.(((....))).)))))...))))))))))(((.(((....((((....)))).))).))).)))))))) ( -40.00) >consensus CCAGCAUGCUCUUACCUGUGUGUGAGUUGGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACUUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGU ...((((((.......((((((((....((...(((((.(((....))).)))))...)))))))))).((((((....((((....)))).))))))..)))))).. (-36.22 = -35.90 + -0.32)

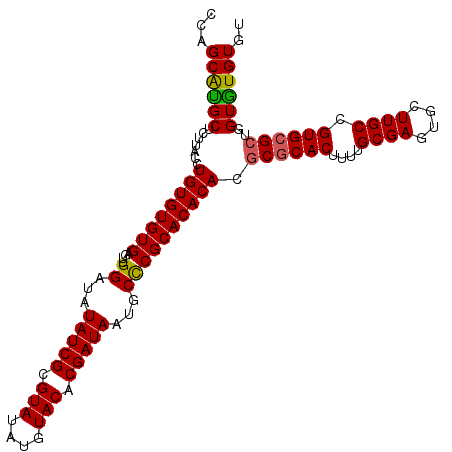

| Location | 10,334,875 – 10,334,983 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -34.52 |
| Energy contribution | -34.64 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334875 108 - 22407834 ACACACACCAGCGCACGGCAAGCACUCGCAAAAGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUUG ......(((.((((((.((........))....))))))(((((((..((...(((((((((....)))))))))...))....))))))).)))..(((....))). ( -41.00) >DroSec_CAF1 736 108 - 1 ACACACACCAGCGCACGGCAAGUACUCGCAAAUGUGCGCGUGUGUGCGAGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGG .......(((((((((.((........))....))))((.((((((.(((...(((((((((....))))))))).......))).)))))).))........))))) ( -39.50) >DroSim_CAF1 736 108 - 1 ACACACACCAGCGCACGGCAAGCACUCGCAAAUGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGG ......(((.(((((((....((....))...)))))))(((((((..((...(((((((((....)))))))))...))....))))))).)))............. ( -40.50) >DroEre_CAF1 427 108 - 1 ACACAUACCACCGCACGGCAAGCACUCGCAAAAGUGCGCUUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACACACACACAGGUAUGCGCACGCUGG .(.((((((..(((((.((........))....)))))..((((((..((...(((((((((....)))))))))...))..))))))....)))))).)........ ( -39.60) >DroYak_CAF1 721 108 - 1 ACACACACUAGCGCACGGCAAGCACUCGCAGAAGUGUGCUUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAUGUGCGCGCUUG ..........(((((((....(((((......)))))(((((((((.(((...(((((((((....))))))))).......))).))))))))).)))))))..... ( -41.90) >consensus ACACACACCAGCGCACGGCAAGCACUCGCAAAAGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCCAACUCACACACAGGUAAGAGCAUGCUGG .......((((((....((..((((........))))((.((((((.(((...(((((((((....))))))))).......))).)))))).))....)).)))))) (-34.52 = -34.64 + 0.12)

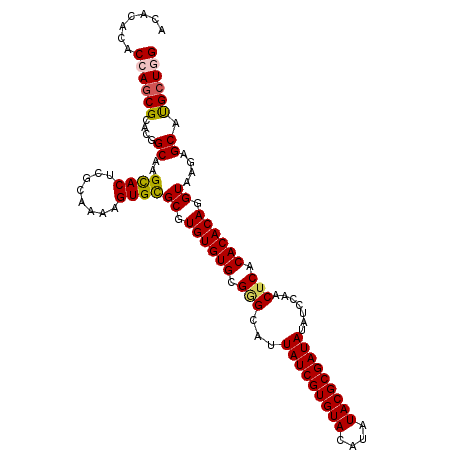
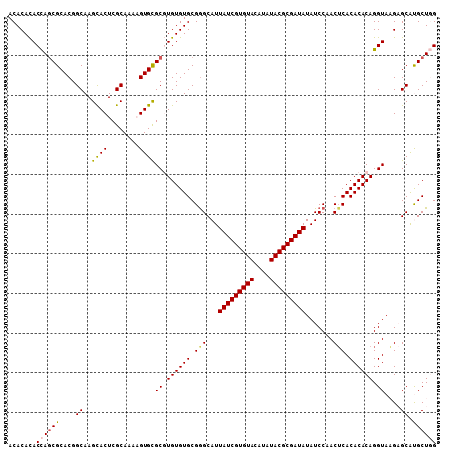
| Location | 10,334,903 – 10,335,023 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -32.88 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334903 120 + 22407834 GGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACUUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((...(((((.(((....))).)))))...)).((((((((((((((....((((....)))).))))))..))))))))..(((((...(((..(((.......)))..))).))))). ( -37.50) >DroSec_CAF1 764 120 + 1 GGAUAUAUCGCGUAUAUGUACACGAUAAUGCUCGCACACACGCGCACAUUUGCGAGUACUUGCCGUGCGCUGGUGUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((((((((((.(((....))).)))))......((((((((((((((....((((....)))).))))))..)))))))))))))..........(((.......)))............ ( -36.10) >DroSim_CAF1 764 120 + 1 GGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACAUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((...(((((.(((....))).)))))...)).((((((((((((((....((((....)))).))))))..))))))))..(((((...(((..(((.......)))..))).))))). ( -37.50) >DroEre_CAF1 455 120 + 1 GGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACAAGCGCACUUUUGCGAGUGCUUGCCGUGCGGUGGUAUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((...(((((.(((....))).)))))...))(((((.(((((((.(......).)))))))..)))))(((((...(((.(((.....))).)))..)))))................. ( -37.20) >DroYak_CAF1 749 120 + 1 GGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACAAGCACACUUCUGCGAGUGCUUGCCGUGCGCUAGUGUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((...(((((.(((....))).)))))...))(((((.(((((((.(......).)))))))..)))))...((((((((((((.....)))...)))).)))))............... ( -36.30) >consensus GGAUAUAUCGCGUAUAUGUACACGAUAAUGCCCGCACACACGCGCACUUUUGCGAGUGCUUGCCGUGCGCUGGUGUGUGUUAUCUAUGUGAUUACGACACCACACGUCAAAAUAAUAGGA ((...(((((.(((....))).)))))...))(((((((..((((((....((((....)))).))))))..)))))))...(((((...(((..(((.......)))..))).))))). (-32.88 = -33.32 + 0.44)

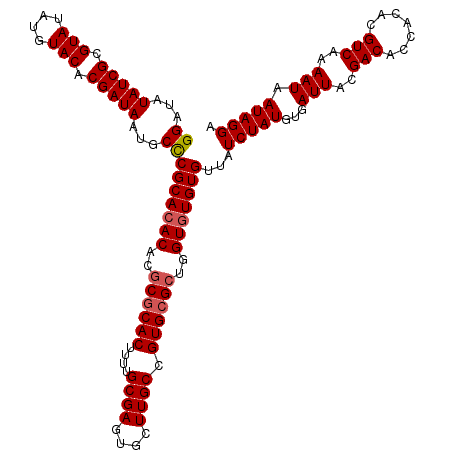
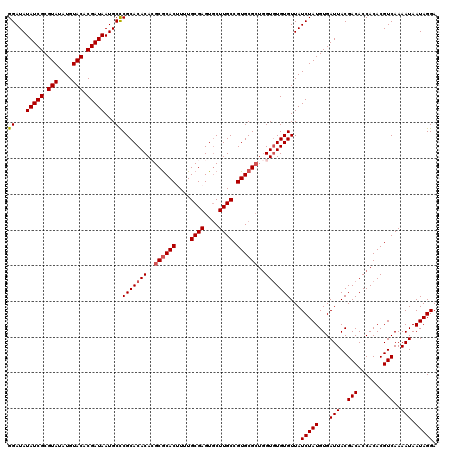
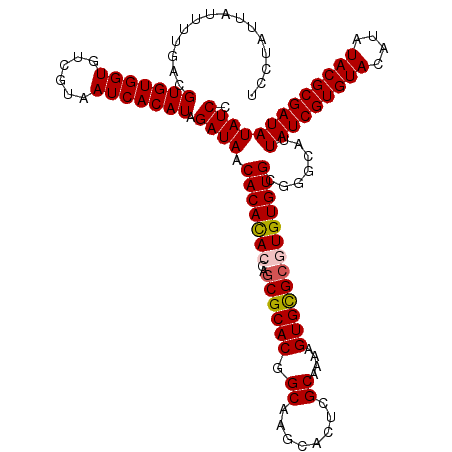
| Location | 10,334,903 – 10,335,023 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -37.76 |
| Energy contribution | -38.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10334903 120 - 22407834 UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACACACCAGCGCACGGCAAGCACUCGCAAAAGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCC .(((.((((((....(((((((......)))))))))))))(((((((..((((((.((........))....))))))))))))).)))...(((((((((....)))))))))..... ( -41.50) >DroSec_CAF1 764 120 - 1 UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACACACCAGCGCACGGCAAGUACUCGCAAAUGUGCGCGUGUGUGCGAGCAUUAUCGUGUACAUAUACGCGAUAUAUCC ...............(((((((......))))))).((((.(((((((..((((((.((........))....))))))))))))).......(((((((((....))))))))))))). ( -40.00) >DroSim_CAF1 764 120 - 1 UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACACACCAGCGCACGGCAAGCACUCGCAAAUGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCC .(((.((((((....(((((((......)))))))))))))(((((((..(((((((....((....))...)))))))))))))).)))...(((((((((....)))))))))..... ( -41.70) >DroEre_CAF1 455 120 - 1 UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACAUACCACCGCACGGCAAGCACUCGCAAAAGUGCGCUUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCC ...............(((((((......))))))).((((..........(((((((.(((((.(.(......).).))))))))))))....(((((((((....))))))))))))). ( -39.70) >DroYak_CAF1 749 120 - 1 UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACACACUAGCGCACGGCAAGCACUCGCAGAAGUGUGCUUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCC ...............(((((.((((...........)))).))))).....((((((.(((((((.(......).)))))))))))))((...(((((((((....)))))))))...)) ( -41.90) >consensus UCCUAUUAUUUUGACGUGUGGUGUCGUAAUCACAUAGAUAACACACACCAGCGCACGGCAAGCACUCGCAAAAGUGCGCGUGUGUGCGGGCAUUAUCGUGUACAUAUACGCGAUAUAUCC ...............(((((((......))))))).((((.(((((((..((((((.((........))....))))))))))))).......(((((((((....))))))))))))). (-37.76 = -38.04 + 0.28)

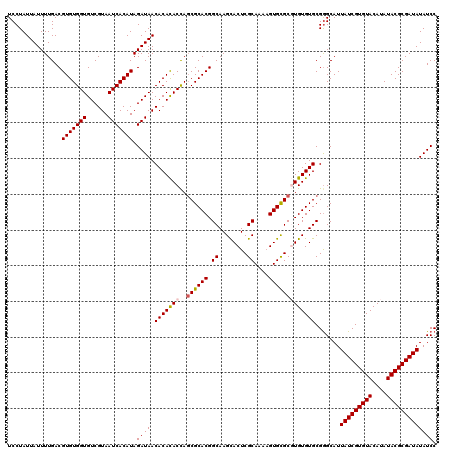
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:01 2006