
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,320,711 – 10,320,861 |
| Length | 150 |
| Max. P | 0.717889 |

| Location | 10,320,711 – 10,320,823 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.14 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10320711 112 + 22407834 GUAUGUAUAUAUUUUGAAAUGUGCUUUUGUUUAUACGAAAGUUUUUUGAGGAUACAUGAAUAUGUACAUACACAUAUGAUGAGUACACUUAUAGCACGUAUCUGUGAAUAUA ((((((((((((((....(((((((((.......((....)).....)))).))))))))))))))))))).((((.((((.((.(.......).)).))))))))...... ( -27.10) >DroSec_CAF1 1606 99 + 1 -----UAUCGAUUUUAAAAUGUGCUUUUGUUUAUACGAAAGUGUUUCGAGGAUACAUACAUAUGUA--------UAUAAUGAGUAUACUUAUAACACGUAUCUGUGAAUAUA -----...............(..(((((((....)))))))..)((((.((((((((((....)))--------)...(((((....))))).....)))))).)))).... ( -20.70) >DroSim_CAF1 1602 99 + 1 -----UAUCGAUUUUAAAAUGUGCUUUUGUUUAUACGAAAGUGUUUCGAGGAUACAUACAUAUGUA--------UAUAAUGAGUAUACUUAUAACACGUAUAUGUGAAUAUA -----.............(((((..((((...((((....))))..))))..))))).((((((((--------(...(((((....))))).....)))))))))...... ( -21.10) >consensus _____UAUCGAUUUUAAAAUGUGCUUUUGUUUAUACGAAAGUGUUUCGAGGAUACAUACAUAUGUA________UAUAAUGAGUAUACUUAUAACACGUAUCUGUGAAUAUA ......................((((((((....))))))))..((((.((((((.......((((........))))(((((....))))).....)))))).)))).... (-14.81 = -14.70 + -0.11)


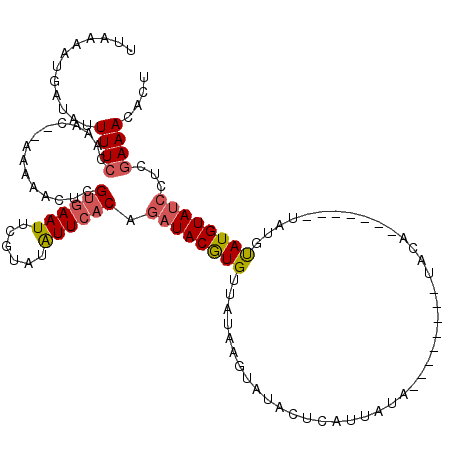
| Location | 10,320,750 – 10,320,861 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -7.93 |
| Energy contribution | -8.29 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10320750 111 - 22407834 UUAAAAUGAUAUUUUCCAAAC--AAAAACCCGUGAAUUCGUAUAUUCACAGAUACGUGCUAUAAGUGUACUCAUCAUAUGUGUAUGUACA-------UAUUCAUGUAUCCUCAAAAAACU ......(((............--........((((((......)))))).((((((((......((((((.(((.....)))...)))))-------)...)))))))).)))....... ( -20.90) >DroSec_CAF1 1640 105 - 1 UUAAAAUGAUAUUUUCCAAAAAAAAAAACUCGUGAAUUCGUAUAUUCACAGAUACGUGUUAUAAGUAUACUCAUUAUA--------UACA-------UAUGUAUGUAUCCUCGAAACACU ............((((...............((((((......)))))).(((((((((.(((.(((((.......))--------))).-------))))))))))))...)))).... ( -16.10) >DroSim_CAF1 1636 103 - 1 UUAAAAUAAUAUUUUCCAAAC--AAAAACUCGUGAAUUCGUAUAUUCACAUAUACGUGUUAUAAGUAUACUCAUUAUA--------UACA-------UAUGUAUGUAUCCUCGAAACACU ............((((.....--....(((..(((...(((((((....)))))))..)))..)))........((((--------(((.-------...))))))).....)))).... ( -15.60) >DroEre_CAF1 1630 99 - 1 UUUGAAUGAAAUUUUCUAAAC--CAAAACUCGUGAAUUCGUAUGUUCACAGAUACAUGUUAUAAGUAUACUCAUCUUA--------UU-----------UAUAUGUAUCUUCGAAACACU ((((((...............--........((((((......)))))).(((((((((.(((((.........))))--------).-----------.)))))))))))))))..... ( -21.00) >DroYak_CAF1 1638 110 - 1 UUAAAAUGAUAUUUUCUAAGC--GAAAACUAGGAAAUUCGCAUGUUCACAGAUACAUGUCAUAAGUAUACUCAAUAUA--------UAUAUUUAUUUUAUGUAUGUAACCUCGAAACACU ............((((...((--(((..........)))))........(.(((((((..(((((((((.........--------)))))))))..))))))).)......)))).... ( -16.80) >consensus UUAAAAUGAUAUUUUCCAAAC__AAAAACUCGUGAAUUCGUAUAUUCACAGAUACGUGUUAUAAGUAUACUCAUUAUA________UACA_______UAUGUAUGUAUCCUCGAAACACU ............((((...............((((((......)))))).((((((((...........................................))))))))...)))).... ( -7.93 = -8.29 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:51 2006