| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,317,954 – 10,318,070 |
| Length | 116 |
| Max. P | 0.955601 |

| Location | 10,317,954 – 10,318,070 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
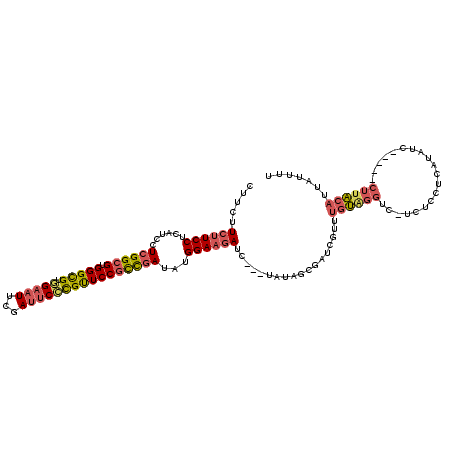
>2L_DroMel_CAF1 10317954 116 + 22407834 CUUCUUCUUCCUCAUCCUCGGCGUGGGCGUGGAAUUCGAUUCCCGUUCCGCCGAUAGGGAAGAUC---UAUAGCGAUCGUUUGUGGGUC-UCUCCUCAUAUCUUAAUCUUACAUUAUUUU .................((((((.(((((.(((((...)))))))))))))))).(((..(((((---((((((....).)))))))))-)..)))........................ ( -36.40) >DroSec_CAF1 3703 116 + 1 CUUCUUCUUCCUCAUCCUCGGCGUGGGCGUGGAAUUCGAUUCCCGUUCCGCCGAUAUGGAAGAUC---UAUUGCGAUCGUUUGUAGGUC-UCUCCUCAUACCUUAAUCUUACAUUAUUUU .................((((((.(((((.(((((...))))))))))))))))...((((((((---(((.((....))..)))))))-).)))......................... ( -33.90) >DroSim_CAF1 2483 116 + 1 CUUCUUCUUCCUCAUCCUCGGCGUGGGCGUGGAAUUCGAUUCCCGUUCCGCCGAUAUGGAAGAUC---UAUAGCGAUCGUUUGUAGGUC-UCUCCUCAUAUCUUAAUCUUACAUUAUUGU .................((((((.(((((.(((((...))))))))))))))))...((((((((---((((((....).)))))))))-).)))......................... ( -34.40) >DroEre_CAF1 2643 110 + 1 CUUCUUCUUCCUCAUCCUCGGCGUGGGCGUAGAAUUUGAUUCCCGUUCCGUCGAUAUGGAAGAUC---UGUUGCGAUCGUUUGUGGGUC-UCUCACCAUAU------CUUACAUUAUUUU .....((((((..(((..(((..((((.((........)).))))..)))..)))..))))))..---..........((.(((((...-.....))))).------...))........ ( -27.80) >DroYak_CAF1 2589 110 + 1 CUUCUUCUUCCGCAUCCUCGGCGUGGGCGUGGAAUUUGAUUCCCGUUCCGCCGAUAUGGAAGAUC---UAUAGCGUUCGCUUGUGGGUC-UCUCACGAUAU------CUUACAUUAUUUU .............(((.((((((.(((((.(((((...))))))))))))))))..((..(((((---((((((....)).))))))))-)..)).)))..------............. ( -36.50) >DroAna_CAF1 2101 110 + 1 UUUCUUCUUCCUCAUCCUAGGUGUGGGUGUAGCAUUGGAGUCUUGCUCCUCUGACAUGGAUAAUGCCAUAAUGCGACCGAAACCACACCGACUCCGAAUAAC------UGGC----CUUU ...................(((((((.((..(((((((((.....))))......((((......)))))))))...))...)))))))....(((......------))).----.... ( -28.20) >consensus CUUCUUCUUCCUCAUCCUCGGCGUGGGCGUGGAAUUCGAUUCCCGUUCCGCCGAUAUGGAAGAUC___UAUAGCGAUCGUUUGUAGGUC_UCUCCUCAUAUC_____CUUACAUUAUUUU .....((((((......((((((.(((((.(((((...))))))))))))))))...))))))..................((((((....................))))))....... (-22.67 = -23.32 + 0.64)


| Location | 10,317,954 – 10,318,070 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
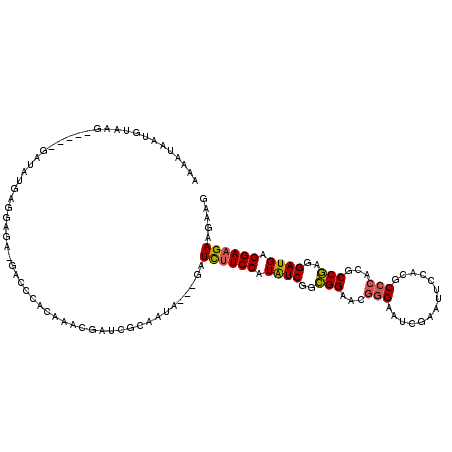
>2L_DroMel_CAF1 10317954 116 - 22407834 AAAAUAAUGUAAGAUUAAGAUAUGAGGAGA-GACCCACAAACGAUCGCUAUA---GAUCUUCCCUAUCGGCGGAACGGGAAUCGAAUUCCACGCCCACGCCGAGGAUGAGGAAGAAGAAG .......((((.((((......((.((...-..)))).....))))..))))---..(((((((..(((((((..(((((((...))))).))..).))))))....).))))))..... ( -28.80) >DroSec_CAF1 3703 116 - 1 AAAAUAAUGUAAGAUUAAGGUAUGAGGAGA-GACCUACAAACGAUCGCAAUA---GAUCUUCCAUAUCGGCGGAACGGGAAUCGAAUUCCACGCCCACGCCGAGGAUGAGGAAGAAGAAG ............((((...((.(((((...-..))).)).))))))......---..((((((...(((((((..(((((((...))))).))..).))))))......))))))..... ( -29.20) >DroSim_CAF1 2483 116 - 1 ACAAUAAUGUAAGAUUAAGAUAUGAGGAGA-GACCUACAAACGAUCGCUAUA---GAUCUUCCAUAUCGGCGGAACGGGAAUCGAAUUCCACGCCCACGCCGAGGAUGAGGAAGAAGAAG .......((((.((((........(((...-..)))......))))..))))---..((((((...(((((((..(((((((...))))).))..).))))))......))))))..... ( -28.54) >DroEre_CAF1 2643 110 - 1 AAAAUAAUGUAAG------AUAUGGUGAGA-GACCCACAAACGAUCGCAACA---GAUCUUCCAUAUCGACGGAACGGGAAUCAAAUUCUACGCCCACGCCGAGGAUGAGGAAGAAGAAG .......(((..(------((...(((...-....))).....)))...)))---..((((((.((((..(((...(((..............)))...)))..)))).))))))..... ( -24.44) >DroYak_CAF1 2589 110 - 1 AAAAUAAUGUAAG------AUAUCGUGAGA-GACCCACAAGCGAACGCUAUA---GAUCUUCCAUAUCGGCGGAACGGGAAUCAAAUUCCACGCCCACGCCGAGGAUGCGGAAGAAGAAG .......(((...------...((....))-.....)))(((....)))...---..((((((...(((((((..(((((((...))))).))..).))))))......))))))..... ( -30.00) >DroAna_CAF1 2101 110 - 1 AAAG----GCCA------GUUAUUCGGAGUCGGUGUGGUUUCGGUCGCAUUAUGGCAUUAUCCAUGUCAGAGGAGCAAGACUCCAAUGCUACACCCACACCUAGGAUGAGGAAGAAGAAA ....----.((.------.((((((......(((((((.....((.(((((.((((((.....))))))..((((.....))))))))).))..)))))))..))))))))......... ( -31.00) >consensus AAAAUAAUGUAAG_____GAUAUGAGGAGA_GACCCACAAACGAUCGCAAUA___GAUCUUCCAUAUCGGCGGAACGGGAAUCGAAUUCCACGCCCACGCCGAGGAUGAGGAAGAAGAAG .........................................................((((((.((((..(((...(((..............)))...)))..)))).))))))..... (-18.58 = -18.52 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:48 2006