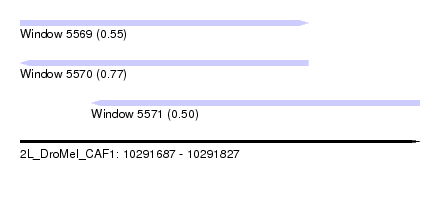
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,291,687 – 10,291,827 |
| Length | 140 |
| Max. P | 0.768624 |

| Location | 10,291,687 – 10,291,788 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -16.19 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10291687 101 + 22407834 UUGAAAAUCAACAACAUAAGCAUAUGUACAUCGAUAUGUACCGAUAUACACAUAC----ACAUAUGCAUGUAUGUAAUCACUUUUUCAAUUACACUUGAUACUUU (((((((......(((((.((((((((.......(((((....))))).......----))))))))...))))).......)))))))................ ( -17.46) >DroSim_CAF1 6079 105 + 1 UUGAAAAUCAACAACAUAAGCAUAUGUACAUCGAUAUGUACCGAUAUCCAAGUACACAUACAUAUGCACGUAUAUAAUCACUUUUUCAAUUACACUUGAUACAUU (((((((............(((((((((.((((........))))......(....).)))))))))..((........)).)))))))................ ( -15.00) >DroYak_CAF1 9294 105 + 1 UUGAAAAUCAACAAAAUAAACAUAUGUACAUCGAUAUGUACCGAUAUACACGUACCCAUACAUGUGCAUGUAUGUAAUCACUUUUUUAAUUUCACUUGAUACUUU .(((((...................((((((....))))))..(((((((.((((........)))).)))))))..............)))))........... ( -16.10) >consensus UUGAAAAUCAACAACAUAAGCAUAUGUACAUCGAUAUGUACCGAUAUACACGUAC_CAUACAUAUGCAUGUAUGUAAUCACUUUUUCAAUUACACUUGAUACUUU (((((((........(((.(((((((((.....((.((((......)))).)).....)))))))))...))).........)))))))................ (-11.92 = -12.26 + 0.34)


| Location | 10,291,687 – 10,291,788 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.83 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10291687 101 - 22407834 AAAGUAUCAAGUGUAAUUGAAAAAGUGAUUACAUACAUGCAUAUGU----GUAUGUGUAUAUCGGUACAUAUCGAUGUACAUAUGCUUAUGUUGUUGAUUUUCAA ...(.(((((((((((((........)))))))).((.(((((...----((((((((((((((((....)))))))))))))))).))))))))))))...).. ( -31.90) >DroSim_CAF1 6079 105 - 1 AAUGUAUCAAGUGUAAUUGAAAAAGUGAUUAUAUACGUGCAUAUGUAUGUGUACUUGGAUAUCGGUACAUAUCGAUGUACAUAUGCUUAUGUUGUUGAUUUUCAA ..((.(((((((((((((........))))))))....(((((.((((((((((.(.(((((......))))).).)))))))))).)))))..)))))...)). ( -28.10) >DroYak_CAF1 9294 105 - 1 AAAGUAUCAAGUGAAAUUAAAAAAGUGAUUACAUACAUGCACAUGUAUGGGUACGUGUAUAUCGGUACAUAUCGAUGUACAUAUGUUUAUUUUGUUGAUUUUCAA ...........(((((((((.(((((((...(((((((....))))))).....((((((((((((....))))))))))))....))))))).)))).))))). ( -28.30) >consensus AAAGUAUCAAGUGUAAUUGAAAAAGUGAUUACAUACAUGCAUAUGUAUG_GUACGUGUAUAUCGGUACAUAUCGAUGUACAUAUGCUUAUGUUGUUGAUUUUCAA ...(.((((((((((((((......))))))))).(((((....))))).(((.((((((((((((....)))))))))))).)))........)))))...).. (-19.60 = -20.83 + 1.23)

| Location | 10,291,712 – 10,291,827 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10291712 115 - 22407834 GUAAAACGGGAUACCCAUCG-UUAUUAUCAUGUGCAAGAAAAAGUAUCAAGUGUAAUUGAAAAAGUGAUUACAUACAUGCAUAUGU----GUAUGUGUAUAUCGGUACAUAUCGAUGUAC .......((....))(((((-.(((((((((((((...............((((((((........))))))))(((((((....)----)))))))))))).)))).))).)))))... ( -27.50) >DroSim_CAF1 6104 120 - 1 GUACAACGAGACACCCAUCGGUUAUUAUCAUGUGCAAGAAAAUGUAUCAAGUGUAAUUGAAAAAGUGAUUAUAUACGUGCAUAUGUAUGUGUACUUGGAUAUCGGUACAUAUCGAUGUAC ((((..((((.(((...((((((((......(((((......))))).....))))))))....)))....(((((((((....)))))))))))))...((((((....)))))))))) ( -29.10) >DroYak_CAF1 9319 119 - 1 GUAAAACGGGACAUCCUUCG-UUUUUAUCAUUUGCAAGAAAAAGUAUCAAGUGAAAUUAAAAAAGUGAUUACAUACAUGCACAUGUAUGGGUACGUGUAUAUCGGUACAUAUCGAUGUAC ..((((((((......))))-))))..(((((((.............)))))))..........(((....(((((((....)))))))..)))..((((((((((....)))))))))) ( -26.72) >consensus GUAAAACGGGACACCCAUCG_UUAUUAUCAUGUGCAAGAAAAAGUAUCAAGUGUAAUUGAAAAAGUGAUUACAUACAUGCAUAUGUAUG_GUACGUGUAUAUCGGUACAUAUCGAUGUAC (((...(((........)))........((((((((.((.......))..(((((((((......)))))))))...))))))))......)))..((((((((((....)))))))))) (-20.38 = -20.50 + 0.12)

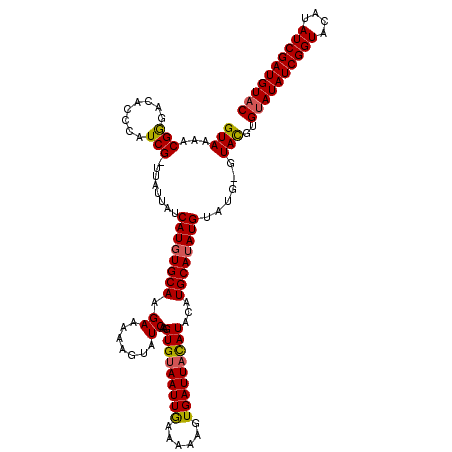
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:40 2006