| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,287,854 – 10,287,956 |
| Length | 102 |
| Max. P | 0.933264 |

| Location | 10,287,854 – 10,287,956 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.09 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
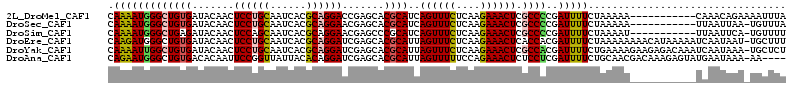
>2L_DroMel_CAF1 10287854 102 + 22407834 CAAAAUGGGCUGUGAUACAACUCCUGCAAUCACGCAGGACCGAGCACGCAUCAGUUUCUCAAGAAACUCGCCCCGAUUUUCUAAAAA-----------CAAACAGAAAAUUUA ......((((.(((.......((((((......)))))).....))).....((((((....)))))).)))).((((((((.....-----------.....)))))))).. ( -27.60) >DroSec_CAF1 502 101 + 1 CAAAAUGGGCUGUGAUACAACUCCUGCAAUCACGCAGGAACGAGCACGCAUCAGUUUCUCAAGAAACUCGCCCCGAUUUUCUAAAAA-----------UUAAUUAA-UGUUUA ......((((.(((.......((((((......)))))).....))).....((((((....)))))).))))..............-----------........-...... ( -24.30) >DroSim_CAF1 2215 101 + 1 CAAAAUGGGCUGAGAUACAACUCCAGCAAUCACGCAGGAACGAGCCCGCAUCAGUUUCUCAAGAAACUCGCCCCGAUUUUCUAAAAU-----------UUAAUUCA-UGUUUU ......((((((((......)))..((......)).......)))))((((.((((.....(((((.(((...))).))))).....-----------..)))).)-)))... ( -18.30) >DroEre_CAF1 5550 112 + 1 CAAGAUGGGCUGUGAUACAACUCCUGCAAUCACGCAGGAUCGAGCACGCAUUAGUUUCUCAAGAAACUCACCACGAUUUUCUAAAAAAAACAUAAAAAUCAAUAAU-UGCUUU .....(((((.(((.......((((((......)))))).....)))))...((((((....))))))..))).((((((..............))))))......-...... ( -22.94) >DroYak_CAF1 5503 112 + 1 CAAAAUUGGCUGUGAUACAACUCCUGCAAUCACGCAGGAUCGAGCACGCAUUAGUUUCUCAAGAAACUCGCCACGAUUUUCUGAAAAGAAGAGACAAAUCAAUAAA-UGCUCU .....................((((((......))))))..(((((.((...((((((....)))))).)).....((((((....))))))..............-))))). ( -27.70) >DroAna_CAF1 5963 108 + 1 CAGAAUGGGCUGUGACACAAUUCCGGUUAUUACACAGGAUCGAGCACGCAUUAGUUUUUCCAGAAACUCUCCUCGAUUUUCUGCAACGACAAAGAGUAUGAAUAAA-AA---- (((......))).....((((((..((..((.((.(((((((((........((((((....))))))...))))))))).)).))..))...)))).))......-..---- ( -16.80) >consensus CAAAAUGGGCUGUGAUACAACUCCUGCAAUCACGCAGGAUCGAGCACGCAUCAGUUUCUCAAGAAACUCGCCCCGAUUUUCUAAAAA___________UCAAUAAA_UGUUUU .(((((((((((((.......((((((......)))))).......))))..((((((....)))))).))))..)))))................................. (-18.09 = -18.09 + 0.00)
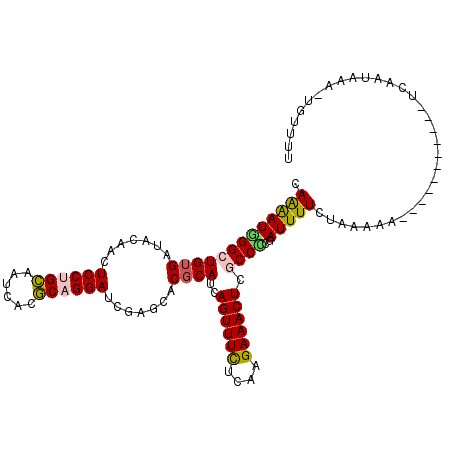
| Location | 10,287,854 – 10,287,956 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10287854 102 - 22407834 UAAAUUUUCUGUUUG-----------UUUUUAGAAAAUCGGGGCGAGUUUCUUGAGAAACUGAUGCGUGCUCGGUCCUGCGUGAUUGCAGGAGUUGUAUCACAGCCCAUUUUG ...((((((((....-----------....))))))))..((((.((((((....))))))((((((.....(.(((((((....))))))).)))))))...))))...... ( -34.60) >DroSec_CAF1 502 101 - 1 UAAACA-UUAAUUAA-----------UUUUUAGAAAAUCGGGGCGAGUUUCUUGAGAAACUGAUGCGUGCUCGUUCCUGCGUGAUUGCAGGAGUUGUAUCACAGCCCAUUUUG ......-........-----------..............((((.((((((....))))))((((((......((((((((....)))))))).))))))...))))...... ( -29.70) >DroSim_CAF1 2215 101 - 1 AAAACA-UGAAUUAA-----------AUUUUAGAAAAUCGGGGCGAGUUUCUUGAGAAACUGAUGCGGGCUCGUUCCUGCGUGAUUGCUGGAGUUGUAUCUCAGCCCAUUUUG ......-........-----------..............((((.((((((....))))))(((((((.....((((.(((....))).)))))))))))...))))...... ( -25.50) >DroEre_CAF1 5550 112 - 1 AAAGCA-AUUAUUGAUUUUUAUGUUUUUUUUAGAAAAUCGUGGUGAGUUUCUUGAGAAACUAAUGCGUGCUCGAUCCUGCGUGAUUGCAGGAGUUGUAUCACAGCCCAUCUUG ...(((-(((((.(((((((............)))))))))))).((((((....))))))..)))(((..((((((((((....))))))).)))...)))........... ( -27.20) >DroYak_CAF1 5503 112 - 1 AGAGCA-UUUAUUGAUUUGUCUCUUCUUUUCAGAAAAUCGUGGCGAGUUUCUUGAGAAACUAAUGCGUGCUCGAUCCUGCGUGAUUGCAGGAGUUGUAUCACAGCCAAUUUUG .(((((-(.(((((..(((((.(((((....))))....).)))))(((((....)))))))))).))))))..(((((((....)))))))((((.....))))........ ( -31.90) >DroAna_CAF1 5963 108 - 1 ----UU-UUUAUUCAUACUCUUUGUCGUUGCAGAAAAUCGAGGAGAGUUUCUGGAAAAACUAAUGCGUGCUCGAUCCUGUGUAAUAACCGGAAUUGUGUCACAGCCCAUUCUG ----..-................((..((.((((((.((.....)).)))))).))..))....((((((.((((((.((......)).)).)))).).))).))........ ( -20.40) >consensus AAAACA_UUUAUUAA___________UUUUUAGAAAAUCGGGGCGAGUUUCUUGAGAAACUAAUGCGUGCUCGAUCCUGCGUGAUUGCAGGAGUUGUAUCACAGCCCAUUUUG ........................................((((.((((((....)))))).....(((..((((((((((....))))))).)))...))).))))...... (-21.23 = -21.82 + 0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:36 2006