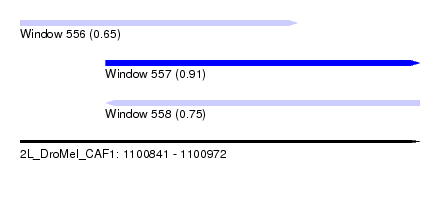
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,100,841 – 1,100,972 |
| Length | 131 |
| Max. P | 0.912673 |

| Location | 1,100,841 – 1,100,932 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
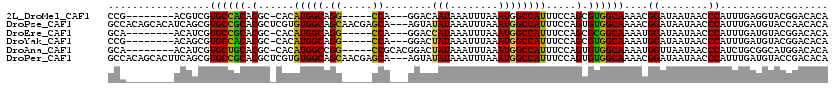
>2L_DroMel_CAF1 1100841 91 + 22407834 -UCG----CACGAUGAUUUAGGCACUUCAUCCU-------CCGACGUCGUGCCACACGCCACAUGGCAGGCCA---GGACAAUAAAUUUAAAUGGCCAUUUCCAGC -..(----(((((((....(((........)))-------....)))))))).....(((....))).(((((---................)))))......... ( -26.69) >DroSec_CAF1 19520 91 + 1 -UCG----CACGAUGAUUUAGGCACCUCAUCCU-------CCGACGUCGUGCCGCACGCCACAUGGCAGGCCA---GGACAAUAAAUUUAAAUGGCCGUUUCCAGC -..(----(((((((....(((........)))-------....)))))))).((..(((....))).(((((---................))))).......)) ( -27.69) >DroSim_CAF1 19285 91 + 1 -UCG----CACGAUGAUUUAGGCACCUCAUCCU-------CCGACGUCGUGCCGCACGCCACAUGGCAGGCCA---GGACAAUAAAUUUAAAUGGCCAUUUCCAGC -..(----(((((((....(((........)))-------....)))))))).((..(((....))).(((((---................))))).......)) ( -27.69) >DroEre_CAF1 22133 95 + 1 -GCGCCAUCACGAUGAUUUGGGUACUUUAUCCU-------GCAACAUCGUGCCGCACGCCACAUGGCAGGCCA---GGACCAUAAAUUUAAAUGGCCAUUUCCAGC -(((((..(((((((....(((((...))))).-------....)))))))......(((....))).)))..---((.((((........)))))).......)) ( -28.80) >DroYak_CAF1 20189 96 + 1 UACACCAUCACUUCAUCAUGGGCACUUCAUCCU-------CCGACAGCGUGCCACACGCCACAUGGCAGGCCA---GGACUAUAAAUUUAAAUGGCCAUUUCCAGC ..................(((((((....((..-------..))....))))).)).(((....))).(((((---................)))))......... ( -20.59) >DroAna_CAF1 20709 105 + 1 -UCGCAAUCAUGAUGACUUAACCACUCCAUCCUUAGCCUUGCAACAUCGUGCUGCACGCCACAUGGCCGGCCGCACGGACUAUAAAUUUAAAUGGCCAUUUCCAGU -..((((....((((............)))).......))))......((((.((..(((....)))..)).))))((.((((........))))))......... ( -22.10) >consensus _UCG____CACGAUGAUUUAGGCACUUCAUCCU_______CCGACAUCGUGCCGCACGCCACAUGGCAGGCCA___GGACAAUAAAUUUAAAUGGCCAUUUCCAGC ....................(((((.......................)))))....(((....))).(((((...(((.......)))...)))))......... (-17.79 = -17.40 + -0.39)

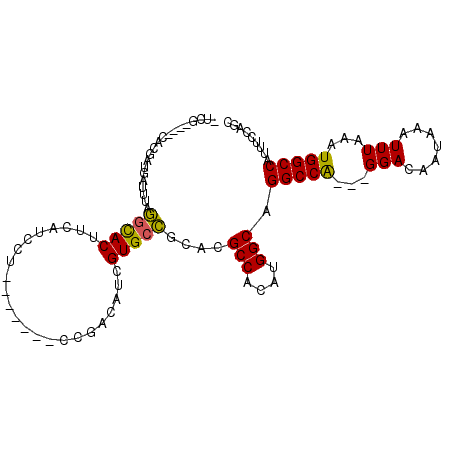
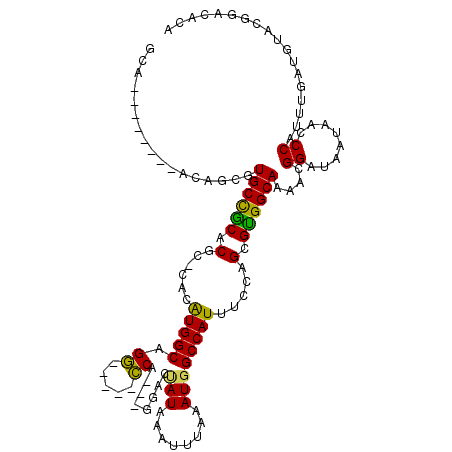
| Location | 1,100,869 – 1,100,972 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -17.41 |
| Energy contribution | -16.30 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1100869 103 + 22407834 CCG--------ACGUCGUGCCACACGC-CACAUGGCAGG-----CCA---GGACAAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAACGGAUAAUAACCCAUUUGAGGUACGGACACA (((--------.(((..((((((.(((-(....))).((-----(((---................))))).......).))))))..))).......((((....).))).)))..... ( -34.59) >DroPse_CAF1 27219 117 + 1 GCCACAGCACAUCAGCGUGCCGCACGCUCGUGUGGCAGCAACGAGCA---AGUAUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAACGGAUAAUAACCCAUUUGAUGUACCAACACA ((....))(((((((..((((((((((((((((....)).)))))).---................(((......)))))))))))....((........))..)))))))......... ( -34.70) >DroEre_CAF1 22165 103 + 1 GCA--------ACAUCGUGCCGCACGC-CACAUGGCAGG-----CCA---GGACCAUAAAUUUAAAUGGCCAUUUCCAGCGCGGCAAAAUGGAUAAUAACCCAUUUGAUGUACGGACACA ...--------(((((.((((((.(((-(....))).((-----(((---................))))).......).))))))((((((........)))))))))))......... ( -35.89) >DroYak_CAF1 20222 103 + 1 CCG--------ACAGCGUGCCACACGC-CACAUGGCAGG-----CCA---GGACUAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAAUGGAUAAUAACCCAUUUGAUGUACGGACACA (((--------(((.(.((((((.(((-(....))).((-----(((---................))))).......).))))))((((((........))))))).))).)))..... ( -34.69) >DroAna_CAF1 20748 106 + 1 GCA--------ACAUCGUGCUGCACGC-CACAUGGCCGG-----CCGCACGGACUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAAUGGUUAAUAACCCAUCUGCGGCAUGGACACA ...--------...((((((((((.((-(((((((..((-----(((...................)))))....))).))))))...((((........)))).))))))))))..... ( -35.81) >DroPer_CAF1 27161 117 + 1 GCCACAGCACUUCAGCGUGCCGCACGCUCGUGUGGCAGCAACGAGCA---AGUAUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAACGGAUAAUAACCCAUUUGAUGUACCGACACA ((((((((.(....((.((((((((....))))))))))...).)).---................(((......))).))))))....((((((.(((.....))).))).)))..... ( -32.20) >consensus GCA________ACAGCGUGCCGCACGC_CACAUGGCAGG_____CCA___GGACUAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAACGGAUAAUAACCCAUUUGAUGUACGGACACA .................((((((.(......(((((.((.....))........(((........)))))))).....).))))))....((........)).................. (-17.41 = -16.30 + -1.11)
| Location | 1,100,869 – 1,100,972 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
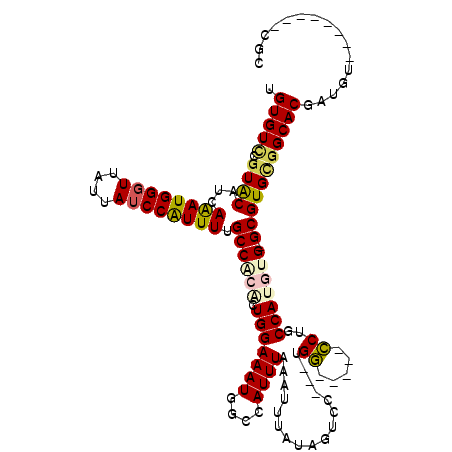
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1100869 103 - 22407834 UGUGUCCGUACCUCAAAUGGGUUAUUAUCCGUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUUGUCC---UGG-----CCUGCCAUGUG-GCGUGUGGCACGACGU--------CGG .....(((.((.((((((((((....))))))))((((((((.......(((((..(((.....)))...---)))-----)).(((....)-)))))))))).)).))--------))) ( -36.80) >DroPse_CAF1 27219 117 - 1 UGUGUUGGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAUACU---UGCUCGUUGCUGCCACACGAGCGUGCGGCACGCUGAUGUGCUGUGGC ......((((((((((((((((....))))))))((((.((((((......)))................---.((((((((....)).))))))))).))))....))))))))..... ( -37.40) >DroEre_CAF1 22165 103 - 1 UGUGUCCGUACAUCAAAUGGGUUAUUAUCCAUUUUGCCGCGCUGGAAAUGGCCAUUUAAAUUUAUGGUCC---UGG-----CCUGCCAUGUG-GCGUGCGGCACGAUGU--------UGC .(((.((((((...((((((((....)))))))).((((((.(((....(((((................---)))-----))..)))))))-))))))))))).....--------... ( -41.59) >DroYak_CAF1 20222 103 - 1 UGUGUCCGUACAUCAAAUGGGUUAUUAUCCAUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUAGUCC---UGG-----CCUGCCAUGUG-GCGUGUGGCACGCUGU--------CGG .((((((((((...((((((((....)))))))).((((((.(((....(((((.(((......)))...---)))-----))..)))))))-)))))))).))))...--------... ( -37.60) >DroAna_CAF1 20748 106 - 1 UGUGUCCAUGCCGCAGAUGGGUUAUUAACCAUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAGUCCGUGCGG-----CCGGCCAUGUG-GCGUGCAGCACGAUGU--------UGC .((((...(((.((((((((........)))))).((((((.(((...(((((.....................))-----))).)))))))-))))))))))).....--------... ( -34.30) >DroPer_CAF1 27161 117 - 1 UGUGUCGGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAUACU---UGCUCGUUGCUGCCACACGAGCGUGCGGCACGCUGAAGUGCUGUGGC .((((.((((....((((((((....))))))))))))))))........(((.................---.((((((((....)).))))))..(((((((......)))))))))) ( -37.40) >consensus UGUGUCCGUACAUCAAAUGGGUUAUUAUCCAUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAGUCC___UGG_____CCUGCCAUGUG_GCGUGCGGCACGAUGU________CGC .(((((.((((...((((((((....)))))))).((((((.(((((((....)))).................((.....))..))))))).)))))))))))................ (-20.25 = -20.33 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:44 2006