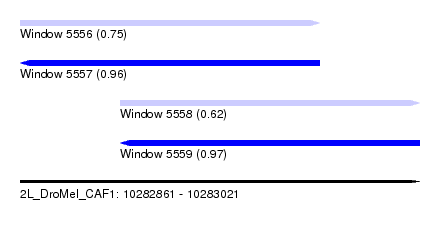
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,282,861 – 10,283,021 |
| Length | 160 |
| Max. P | 0.967163 |

| Location | 10,282,861 – 10,282,981 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -29.30 |
| Energy contribution | -31.97 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282861 120 + 22407834 UACCCCCGAAUGCAUUCGACUUGUUGGAAUCAGUUCGCAGCAAUUUAGUGCAGGUUUCUACUAUAUCAGAGUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACCU ..........((((((.((.(((((((((....))).)))))))).))))))(((........((((((......))))))(((((....))))).(((((((......)))))))))). ( -35.00) >DroSec_CAF1 3070 120 + 1 UACCCCCGAAUGCAUUCGACUUGUUGGAAUCAGUUCGCAGCAGUUUGGUGCAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUU ..........(((((..((((.(((((((....))).))))))))..)))))...........(((((((....)))))))(((((....))))).(((((((......))))))).... ( -36.70) >DroSim_CAF1 2837 120 + 1 UACCCCCGAAUGCAUUCGACUUGUUGGAAUCAGUUCGCAGCAUUUUGGUACAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUU .(((((.((((((...(((.(((.......))).)))..)))))).))....)))........(((((((....)))))))(((((....))))).(((((((......))))))).... ( -33.00) >consensus UACCCCCGAAUGCAUUCGACUUGUUGGAAUCAGUUCGCAGCAAUUUGGUGCAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUU ..........(((((.(((.(((((((((....))).)))))).))))))))(((........(((((((....)))))))(((((....))))).(((((((......)))))))))). (-29.30 = -31.97 + 2.67)


| Location | 10,282,861 – 10,282,981 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -34.52 |
| Energy contribution | -34.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282861 120 - 22407834 AGGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGACUCUGAUAUAGUAGAAACCUGCACUAAAUUGCUGCGAACUGAUUCCAACAAGUCGAAUGCAUUCGGGGGUA ..(((((((((......))))))))).((((.((((((((((((((....))))))...((((....))))...........))).)))))..........(((((....))))))))). ( -35.70) >DroSec_CAF1 3070 120 - 1 AAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGCACCAAACUGCUGCGAACUGAUUCCAACAAGUCGAAUGCAUUCGGGGGUA ..(((((((((......)))))))))(((......)))((((((((....)))))))).((((....))))(((...(((.((((...(((((......)))))..))))..))).))). ( -36.10) >DroSim_CAF1 2837 120 - 1 AAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGUACCAAAAUGCUGCGAACUGAUUCCAACAAGUCGAAUGCAUUCGGGGGUA ..(((((((((......))))))))).(((((((.(..((((((((....))))))))..).))))......((..(((((..(((..((.......))..)))...)))))..))))). ( -38.00) >consensus AAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGCACCAAAAUGCUGCGAACUGAUUCCAACAAGUCGAAUGCAUUCGGGGGUA ..(((((((((......))))))))).(((((((.(..((((((((....))))))))..).))))......((....(((..(((..((.......))..)))...)))....))))). (-34.52 = -34.30 + -0.22)

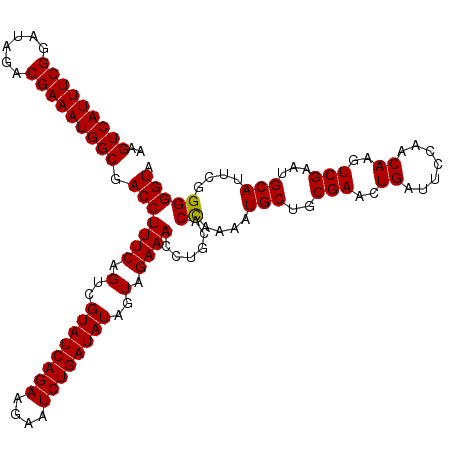
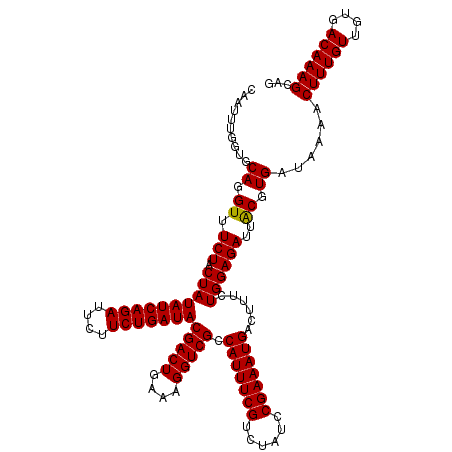
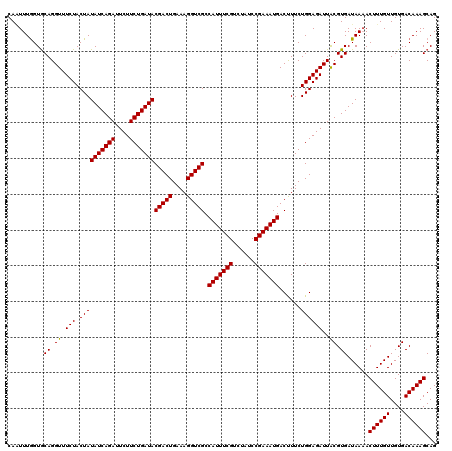
| Location | 10,282,901 – 10,283,021 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -28.49 |
| Energy contribution | -30.27 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282901 120 + 22407834 CAAUUUAGUGCAGGUUUCUACUAUAUCAGAGUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACCUUCUGGAGAUUACGUGAUAACACUUUGUUGUGACAAAGCAG ........(((.((((((((...((((((......))))))(((((....))))).(((((((......)))))))......))))))))..((.(((((.....))))).))...))). ( -34.50) >DroSec_CAF1 3110 120 + 1 CAGUUUGGUGCAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUUUCUGGAGAUUGCGUGAUAAAACUUUGUUGUGACAAAGCAG ..((((...((((.((((((...(((((((....)))))))(((((....))))).(((((((......)))))))......))))))))))((.((((.......)))).)).)))).. ( -33.20) >DroSim_CAF1 2877 120 + 1 CAUUUUGGUACAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUUUCUGGAGAUUACGUGAUAAAACUUUGUUGUGACAAAGCAG ..(((((.(((.((((((((...(((((((....)))))))(((((....))))).(((((((......)))))))......))))))))..))).)))))((((((....))))))... ( -32.10) >consensus CAAUUUGGUGCAGGUUUCUACUAUAUCAGAUUCUUCUGAUACGACUGAAAGGUCGCCAUUUCGUCUAUCCGAAAUGACUUUCUGGAGAUUACGUGAUAAAACUUUGUUGUGACAAAGCAG ..........((.((.(((.((((((((((....)))))))(((((....))))).(((((((......)))))))......))))))..)).))......((((((....))))))... (-28.49 = -30.27 + 1.78)

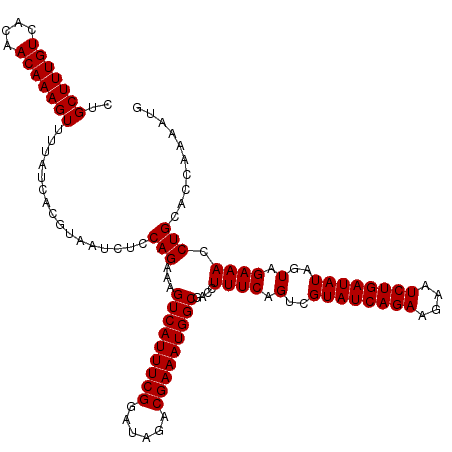
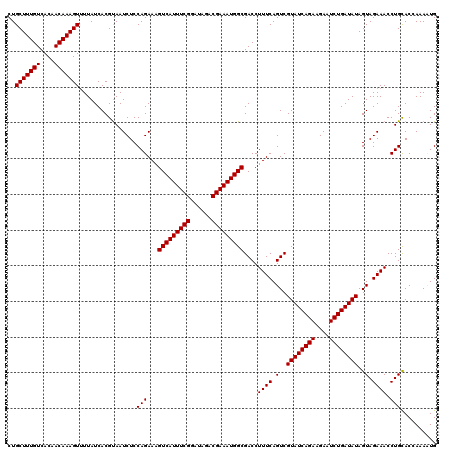
| Location | 10,282,901 – 10,283,021 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -30.17 |
| Energy contribution | -30.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282901 120 - 22407834 CUGCUUUGUCACAACAAAGUGUUAUCACGUAAUCUCCAGAAGGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGACUCUGAUAUAGUAGAAACCUGCACUAAAUUG ..(((((((....)))))))......................(((((((((......)))))))))(((......)))((((((((....)))))))).((((....))))......... ( -32.00) >DroSec_CAF1 3110 120 - 1 CUGCUUUGUCACAACAAAGUUUUAUCACGCAAUCUCCAGAAAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGCACCAAACUG ..(((((((....)))))))................(((...(((((((((......)))))))))(((......)))((((((((....)))))))).((((....))))......))) ( -32.10) >DroSim_CAF1 2877 120 - 1 CUGCUUUGUCACAACAAAGUUUUAUCACGUAAUCUCCAGAAAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGUACCAAAAUG ..(((((((....)))))))................(((...(((((((((......)))))))))....((((.(..((((((((....))))))))..).)))).))).......... ( -30.10) >consensus CUGCUUUGUCACAACAAAGUUUUAUCACGUAAUCUCCAGAAAGUCAUUUCGGAUAGACGAAAUGGCGACCUUUCAGUCGUAUCAGAAGAAUCUGAUAUAGUAGAAACCUGCACCAAAAUG ..(((((((....)))))))................(((...(((((((((......)))))))))....((((.(..((((((((....))))))))..).)))).))).......... (-30.17 = -30.17 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:28 2006