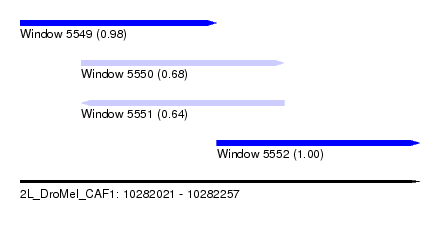
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,282,021 – 10,282,257 |
| Length | 236 |
| Max. P | 0.999246 |

| Location | 10,282,021 – 10,282,137 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 98.10 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -37.38 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282021 116 + 22407834 UUGGUUGCUGUUAACAAGCGCUCCUUCAUGAUCUCUUCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU ..((((((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....)))))...))).. ( -37.10) >DroSec_CAF1 2232 116 + 1 UUGGUUGCUGUUAACAAGCGCUCCUUCAUGAUCUCUUCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU ..((((((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....)))))...))).. ( -37.10) >DroSim_CAF1 1995 116 + 1 UUGGUUGCUGUUAACAAGCGCUCCUUCAUGAUCUCUUCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU ..((((((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....)))))...))).. ( -37.10) >DroEre_CAF1 2240 116 + 1 UUAGUUGCUGUUAACAAGCGCUCCUUCAUGAUCUCUUCAGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCAUGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU .(((.(((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....))))).))).... ( -37.90) >DroYak_CAF1 1913 116 + 1 UUAGUUGCUGUUAACAAGCGCUCUUUCAUGAUCUCUUCAGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCAUGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU .(((.(((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....))))).))).... ( -35.10) >consensus UUGGUUGCUGUUAACAAGCGCUCCUUCAUGAUCUCUUCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUU .(((.(((((........(((((((...(((.....)))...))).)).))(((((((....((.(((((((....)))))))..))....))))))).....))))).))).... (-37.38 = -36.74 + -0.64)


| Location | 10,282,057 – 10,282,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -38.28 |
| Energy contribution | -38.64 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282057 120 + 22407834 UCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUUCGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCG .(((((((.((....(((((((....((.(((((((....)))))))..))....)))))))...(((..(((((.((.....(((....)))....)))))))..))))).)))).))) ( -42.70) >DroSec_CAF1 2268 120 + 1 UCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUUCGGACAACUGUCAGUCGGGGAUGAAGAUUGGCCAGUCCG .(((((((.(((((.(((((((....((.(((((((....)))))))..))....)))))))........(((((.((.....(((....)))....)))))))..))))).)))).))) ( -44.50) >DroSim_CAF1 2031 120 + 1 UCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUUCGGACAACUGUCAGUCGGGGAUGAAGAUUGGCCAGUCCG .(((((((.(((((.(((((((....((.(((((((....)))))))..))....)))))))........(((((.((.....(((....)))....)))))))..))))).)))).))) ( -44.50) >DroEre_CAF1 2276 120 + 1 UCAGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCAUGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUUCGAACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCU ......(((..(...(((((((....((.(((((((....)))))))..))....)))))))...((((((.((..(((..(..((....))..)..)))...))..)))))))..))). ( -37.60) >DroYak_CAF1 1949 120 + 1 UCAGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCAUGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUACGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCU ......(((..(...(((((((....((.(((((((....)))))))..))....)))))))...((((((.((..(((.((.(((....))).)).)))...))..)))))))..))). ( -41.80) >consensus UCGGUUGGGUAGUCGGGAAGCUUUAAGUAGUGAACGCACGUGUUCACGGAUGGAUAGCUUCCAACGCCAGCAUCUACCUUUCGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCG ..((((((.(((((.(((((((....((.(((((((....)))))))..))....)))))))........(((((.((.....(((....)))....)))))))..))))).)))).)). (-38.28 = -38.64 + 0.36)

| Location | 10,282,057 – 10,282,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282057 120 - 22407834 CGGACUGGCCAACCUUCAUCCCCGACUGACAGUUGUCCGAAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACGUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACCGA (((....((((.....((((..((((.....)))).((....))..)))).))))((((((((((........((((((......)))))).......))))).))))).......))). ( -35.66) >DroSec_CAF1 2268 120 - 1 CGGACUGGCCAAUCUUCAUCCCCGACUGACAGUUGUCCGAAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACGUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACCGA (((....((((.....((((..((((.....)))).((....))..)))).))))((((((((((........((((((......)))))).......))))).))))).......))). ( -35.66) >DroSim_CAF1 2031 120 - 1 CGGACUGGCCAAUCUUCAUCCCCGACUGACAGUUGUCCGAAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACGUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACCGA (((....((((.....((((..((((.....)))).((....))..)))).))))((((((((((........((((((......)))))).......))))).))))).......))). ( -35.66) >DroEre_CAF1 2276 120 - 1 AGGACUGGCCAACCUUCAUCCCCGACUGACAGUUGUUCGAAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACAUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACUGA .((....((((.....((((..((((.....)))).((....))..)))).))))((((((((((........((((((......)))))).......))))).)))))...))...... ( -31.66) >DroYak_CAF1 1949 120 - 1 AGGACUGGCCAACCUUCAUCCCCGACUGACAGUUGUCCGUAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACAUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACUGA .((....((((.....((((((..((.(((....))).))..))..)))).))))((((((((((........((((((......)))))).......))))).)))))...))...... ( -34.16) >consensus CGGACUGGCCAACCUUCAUCCCCGACUGACAGUUGUCCGAAAGGUAGAUGCUGGCGUUGGAAGCUAUCCAUCCGUGAACACGUGCGUUCACUACUUAAAGCUUCCCGACUACCCAACCGA .((....((((.....((((..((((.....)))).((....))..)))).))))((((((((((........((((((......)))))).......))))).)))))...))...... (-33.44 = -33.28 + -0.16)

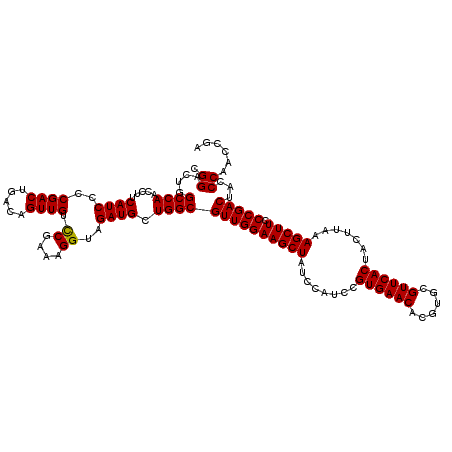
| Location | 10,282,137 – 10,282,257 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -48.66 |
| Consensus MFE | -43.00 |
| Energy contribution | -42.40 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10282137 120 + 22407834 UCGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCGCCUGGCGGCAGGCUGCUGCAACCGGUUGUGAAUUGGAGGAACGCCUUCCUCUCGUCACCCGACAUGCUCAACAAAACGUU .(((((....))).(((((((((((.(((((..(((((.(((....))).)))))...)))))............((((((....))))))))))).)))))).............)).. ( -46.50) >DroSec_CAF1 2348 120 + 1 UCGGACAACUGUCAGUCGGGGAUGAAGAUUGGCCAGUCCGCCUGGUGGCAGGCUGCUGCAACCGGUUGUGAACUGGAGGAACGCCUUCCUCUCGUCUCCCGACAUGCUCAACAAAACGUU .(((((....))).(((((((((((........((((.((((((((.((((....)))).)))))..))).))))((((((....))))))))))).)))))).............)).. ( -47.40) >DroSim_CAF1 2111 120 + 1 UCGGACAACUGUCAGUCGGGGAUGAAGAUUGGCCAGUCCGCCUGGUGGCAGGCUGCUGCAACCGGUUGUGAACUGGAGGAACGCCUUCCUCUCGUCUCCCGACAUGCUCAACAAAACGUU .(((((....))).(((((((((((........((((.((((((((.((((....)))).)))))..))).))))((((((....))))))))))).)))))).............)).. ( -47.40) >DroEre_CAF1 2356 120 + 1 UCGAACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCUCCUGGGGGCAGGCUACUGCAACCAGUUGUGAAUUGGAGGAACGCCUUCCUCUCAUCUCCCGACAUGCUUAACAAAACGUU ..............((((((((((((((..(((...(((((((((..((((....))))..)))..........))))))..)))..))).))))).))))))................. ( -49.20) >DroYak_CAF1 2029 120 + 1 ACGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCUCCUGGGGGCAGGCUACUGCAACCAGUUGUGAAUUGGAGGAACGCCUUCCUUUCGUCUCCCGACAUGCUUAACAAAACGUU ((((((....))).((((((((((((((..(((...(((((((((..((((....))))..)))..........))))))..)))...)))))))).)))))).............))). ( -52.80) >consensus UCGGACAACUGUCAGUCGGGGAUGAAGGUUGGCCAGUCCGCCUGGGGGCAGGCUGCUGCAACCGGUUGUGAAUUGGAGGAACGCCUUCCUCUCGUCUCCCGACAUGCUCAACAAAACGUU .(((((....))).(((((((((((........((((((..((((..((((....))))..))))..).).))))((((((....))))))))))).)))))).............)).. (-43.00 = -42.40 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:21 2006