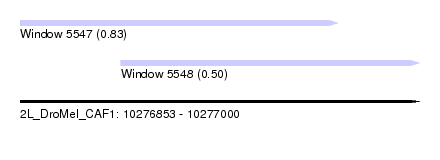
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,276,853 – 10,277,000 |
| Length | 147 |
| Max. P | 0.827495 |

| Location | 10,276,853 – 10,276,970 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.35 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -21.91 |
| Energy contribution | -23.03 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
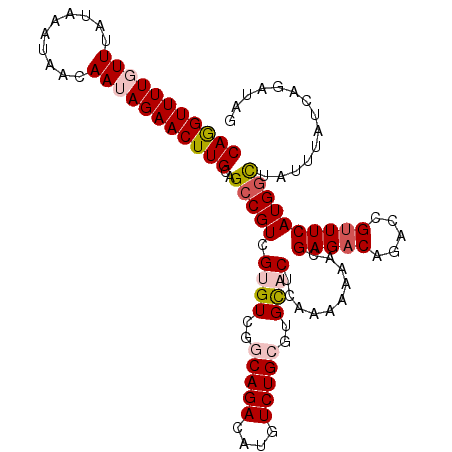
>2L_DroMel_CAF1 10276853 117 + 22407834 CUUAAAUGUUAAUCU---CCUAAAAUAUGCCUGCAAAAUGCAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGA ............(((---(........((..((((...(((((((((((((..........)))))))))))))((.(.((((((....)))))).).)).))))..)).......)))) ( -31.06) >DroSec_CAF1 5010 117 + 1 CUUAAAUGUUAAUCU---CCUAAAAUGUGCCCGCAAAAUGCAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGACAGACAUGUCUGCGUGCACUCAAAAAAACGAGA ............(((---(.......((((.((((...(((((((((((((..........)))))))))))))...(.((((((....)))))).))))).))))..........)))) ( -34.83) >DroSim_CAF1 5148 117 + 1 CUUAAAUGUUAAUCU---CCUAAAAUGUGCCCGAAAAAUGCAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGA ............(((---(.......((((.(......(((((((((((((..........)))))))))))))((.(.((((((....)))))).).))).))))..........)))) ( -31.93) >DroEre_CAF1 5023 120 + 1 CUUAAAUGUUAAUCUGUUGCUAAAAUGUGACCACGAAACACAGGUUUUGUUUAUAAAUAACAAAAGAACUUGCAGCCGUCGAGUCGGCAGACAUGUCUGAGUGUACUCAAAAAAACGAGA ......(((((....((..(......)..)).(((((((....))))))).......)))))......((((..((((......)))).........((((....))))......)))). ( -24.30) >DroYak_CAF1 4964 120 + 1 CCUAAAUGUUAAUCUGUUGCUAAAAUGUGCUCACUAAACGCAAGUUUUGUUUAUAAAUCACAAAAGAACUUGCAGCCGUCGAGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGA ............((((((........((((.(.......(((((((((................))))))))).((((......))))(((....)))..).)))).......))).))) ( -26.25) >consensus CUUAAAUGUUAAUCU___CCUAAAAUGUGCCCGCAAAAUGCAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGA ............(((...........((((.(.......((((((((((((..........)))))))))))).((.(.((((((....)))))).).))).))))...........))) (-21.91 = -23.03 + 1.12)



| Location | 10,276,890 – 10,277,000 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -21.16 |
| Energy contribution | -22.28 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10276890 110 + 22407834 CAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGACAGACCGUUUCAUGGCUUUUUAUCAGAUAG (((((((((((..........))))))))))).((((((.((((..(((((....)))))..))))..........(((((.....)))))))))))............. ( -30.40) >DroSec_CAF1 5047 110 + 1 CAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGACAGACAUGUCUGCGUGCACUCAAAAAAACGAGACAGACCGUUUCAUGGCUAUUUAUCAGAUAG (((((((((((..........))))))))))).((((((.((((((.((((....)))))).))))..........(((((.....)))))))))))............. ( -29.40) >DroSim_CAF1 5185 110 + 1 CAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGACAGACCGUUUCAUGGCUAUUUAUCAGAUAG (((((((((((..........))))))))))).((((((.((((..(((((....)))))..))))..........(((((.....)))))))))))............. ( -31.00) >DroEre_CAF1 5063 110 + 1 CAGGUUUUGUUUAUAAAUAACAAAAGAACUUGCAGCCGUCGAGUCGGCAGACAUGUCUGAGUGUACUCAAAAAAACGAGACAGACUGCUUCAUGGUAUUUUAUUAGAUAU (((((((((((.......))))...)))))))..((((......))))....((((((((.(((.(((........))))))...((((....)))).....)))))))) ( -23.50) >DroYak_CAF1 5004 110 + 1 CAAGUUUUGUUUAUAAAUCACAAAAGAACUUGCAGCCGUCGAGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGACAGACGGUUUCAUGUCAGUUUAUAACAUAG ....((((((.........))))))(((((.(((((((((..(((....))).(((((.(((............))))))))))))))....))).)))))......... ( -27.00) >consensus CAGGUUUUGUUUAUAAAUAACAAUAGAACUUGCAGCCGUCGUGUCGGCAGACAUGUCUGCGUGCACUCAAAAAAACGAGACAGACCGUUUCAUGGCUAUUUAUCAGAUAG (((((((((((..........)))))))))))..(((((.((((..(((((....)))))..))))..........(((((.....)))))))))).............. (-21.16 = -22.28 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:17 2006