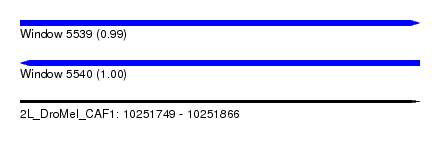
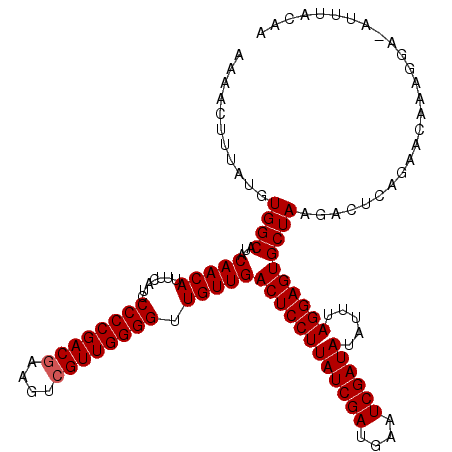
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,251,749 – 10,251,866 |
| Length | 117 |
| Max. P | 0.999991 |

| Location | 10,251,749 – 10,251,866 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -25.55 |
| Energy contribution | -25.79 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10251749 117 + 22407834 GUGUA-AU-UCCUUUGUUCCGAGUCAUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUCGUCGGGGGAUGAAAUGUUGUAUGCCACAUAAAGUUUU .....-..-..((((((.....((.(((((((((((.....((((((....)))))))))))(((....((((.(((....))).))))..)))..)))))))).....)))))).... ( -29.20) >DroSec_CAF1 5790 118 + 1 UUGUAAAU-UCCUUUGUUCCGAGUCUUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUCGUCGGGGGAUGAAAUGUUGUAUGCCACAUAAAGUUUU ((((..((-((((.......((((......)))).......((((((....))))))))))))..))))((((.(((....))).))))......((((.(....).))))........ ( -28.50) >DroSim_CAF1 4205 118 + 1 UUGUAAAU-UCCUUUGUUCUGAGUCUUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUCGUCGGGGGAUGAAAUGUUGUAUGCCACAUAAAGUUUU ........-..((((((..((.((..((((((((((.....((((((....)))))))))))(((....((((.(((....))).))))..)))..)))))...)))).)))))).... ( -29.40) >DroEre_CAF1 4201 119 + 1 UUGUAAAAGAAAUUUUUUCUUAAUAUUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUCGUCGGGGGAUGAAAUGUUGUAUGCCAUAUAAAGUUUU ........((((((((......((((.(((((((((.....((((((....)))))))))))(((....((((.(((....))).))))..)))..)))))))).......)))))))) ( -28.32) >DroYak_CAF1 4173 117 + 1 UUUUAAAU--UCUUUGUUUUGAAUCUUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUUGUCGGGGGAUGAAAUGUUGUAUGCCAUAUAAAGUUUU ........--.((((((..((.....((((((((((.....((((((....))))))))))))...((.((((..((((...)))))))).))....)))).....)).)))))).... ( -27.40) >consensus UUGUAAAU_UCCUUUGUUCUGAGUCUUAGCACUCCUAAAUAUAUCGAUUCAUCGAUAAGGAGUCAACAACCCCAACGACUUCGUCGGGGGAUGAAAUGUUGUAUGCCACAUAAAGUUUU ...........((((((.........((((((((((.....((((((....)))))))))))(((....((((.(((....))).))))..)))..)))))........)))))).... (-25.55 = -25.79 + 0.24)

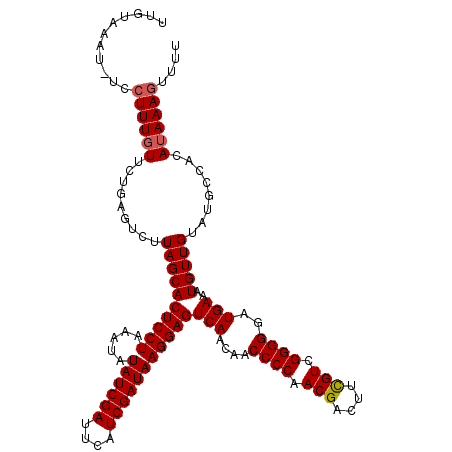
| Location | 10,251,749 – 10,251,866 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.62 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10251749 117 - 22407834 AAAACUUUAUGUGGCAUACAACAUUUCAUCCCCCGACGAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAUGACUCGGAACAAAGGA-AU-UACAC ..........(((((...(((((.......((((((((....)))))))).)))))((((((((((((....)))))).....)))))))))))................-..-..... ( -34.70) >DroSec_CAF1 5790 118 - 1 AAAACUUUAUGUGGCAUACAACAUUUCAUCCCCCGACGAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAAGACUCGGAACAAAGGA-AUUUACAA ...........((((...(((((.......((((((((....)))))))).)))))((((((((((((....)))))).....)))))))))).................-........ ( -33.40) >DroSim_CAF1 4205 118 - 1 AAAACUUUAUGUGGCAUACAACAUUUCAUCCCCCGACGAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAAGACUCAGAACAAAGGA-AUUUACAA ...........((((...(((((.......((((((((....)))))))).)))))((((((((((((....)))))).....)))))))))).................-........ ( -33.40) >DroEre_CAF1 4201 119 - 1 AAAACUUUAUAUGGCAUACAACAUUUCAUCCCCCGACGAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAAUAUUAAGAAAAAAUUUCUUUUACAA ....(((.(((((((...(((((.......((((((((....)))))))).)))))((((((((((((....)))))).....))))))))).)))).))).................. ( -35.40) >DroYak_CAF1 4173 117 - 1 AAAACUUUAUAUGGCAUACAACAUUUCAUCCCCCGACAAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAAGAUUCAAAACAAAGA--AUUUAAAA ...........((((...(((((.......(((((((......))))))).)))))((((((((((((....)))))).....))))))))))((((((........))--)))).... ( -33.40) >consensus AAAACUUUAUGUGGCAUACAACAUUUCAUCCCCCGACGAAGUCGUUGGGGUUGUUGACUCCUUAUCGAUGAAUCGAUAUAUUUAGGAGUGCUAAGACUCAGAACAAAGGA_AUUUACAA ...........((((...(((((.......((((((((....)))))))).)))))((((((((((((....)))))).....)))))))))).......................... (-32.30 = -32.50 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:09 2006