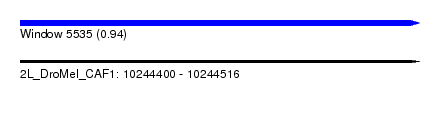
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,244,400 – 10,244,516 |
| Length | 116 |
| Max. P | 0.939515 |

| Location | 10,244,400 – 10,244,516 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -23.29 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10244400 116 + 22407834 AAGCCAAUUGAUUAGCUACUUUUCAUGUUAAAAAAUU---GUUUUUGUGUUUGG-CUUACCAUUUUGUUCGGUGACUUUGCUAAAUAAUUCCGCACUGGCAAAGUCAUCAAUGUUUUCCU (((((((....(((((..........)))))((((..---..))))....))))-)))............(((((((((((((.............)))))))))))))........... ( -26.52) >DroPse_CAF1 204 102 + 1 AA----AUUAGUCGAUUA---------UUAUGAAAUGUGUAUUUUU--AUAUGUAAUUACCAUUUUUUACAGUGGUUGAACUAAG---CUCAGCACUUGCAAAAUCGUCAAUAUUUUCUU ..----.(((((((((((---------((..((((((.(((.((..--......)).)))))))))....)))))))).))))).---....((....)).................... ( -15.30) >DroSec_CAF1 3872 108 + 1 A-----AUUGAUCAGCUACUUUUCAUGUUAAUAAAUG---GUUUUUGUUUUUGC-AUUACCAUUUGGUUCGGUGACUUUGCUAAA---UUCCGCACUGGCAAAGUCAUCAAUGUUUUCCU .-----.........................((((((---((...(((....))-)..))))))))....(((((((((((((..---........)))))))))))))........... ( -27.70) >DroSim_CAF1 3836 113 + 1 AAGCCAAUUGAUCAGCUACUUUUCAUGUUAAUAAAUG---GUUUUUGUUUUUGC-AUUACCAUUUGGUUCGGUGACUUUGCUAAA---CUCCGCACUGACAAAGUCAUCAAUGUUUUCCU .((((.(((((.((...........)))))))(((((---((...(((....))-)..))))))))))).(((((((((((....---.........).))))))))))........... ( -24.02) >DroEre_CAF1 3769 108 + 1 AAACCAAUUAAUUAAUUGCAGUUUUAGUUGAAAUAUG---CUU-----UUAUGC-AUUACCAUUUUGUUCGCUGACUUUGCUAAA---UUCCGCACUGGCAAAGUCAUCAAUGUUUUCCU .......((((((((........))))))))...(((---(..-----....))-)).............(.(((((((((((..---........))))))))))).)........... ( -20.60) >DroYak_CAF1 3695 108 + 1 AAAGCAAUUGAUGAAAUGCAAUUCAUGUACAAAAAUU---CUU-----AUAUGC-AUUACCAUUUGGUUCGGUGACUUUGCUAAA---UUCCGCACUGGCAAAGUCAUCAAUGUUCUCCU ..((((...(((.(((((.(((.((((((........---..)-----))))).-)))..))))).))).(((((((((((((..---........)))))))))))))..))))..... ( -25.60) >consensus AA_CCAAUUGAUCAACUACUUUUCAUGUUAAAAAAUG___GUUUUU__UUAUGC_AUUACCAUUUGGUUCGGUGACUUUGCUAAA___UUCCGCACUGGCAAAGUCAUCAAUGUUUUCCU ......................................................................(((((((((((((.............)))))))))))))........... (-12.36 = -12.97 + 0.61)

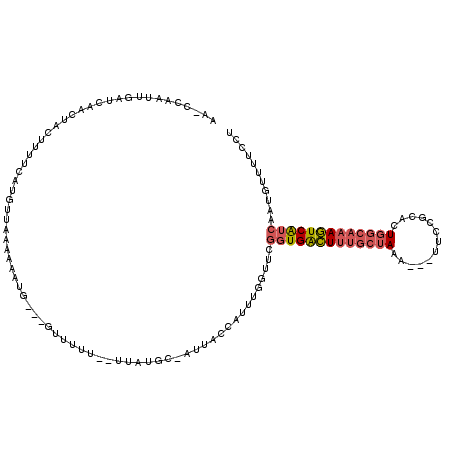
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:04 2006